Dissecting Cleavage Mechanisms
in Tensed Collagen Fibrils
using Reactive Molecular Dynamics Simulations
Introduction
Concerning collagen
An orange a day keeps the scurvy away


Citrus fruits contain Vitamin C ( 🟠 )
1747, HMS Salisbury, James Lind performs the first clinical trial with a control group [1]
Image sources: left: The Capture of Chandernagore, March 1757 by Dominic Serres, the Elder in 1771, oil on canvas.
right: Riftbound, the League of Legends TCG ©Riot Games
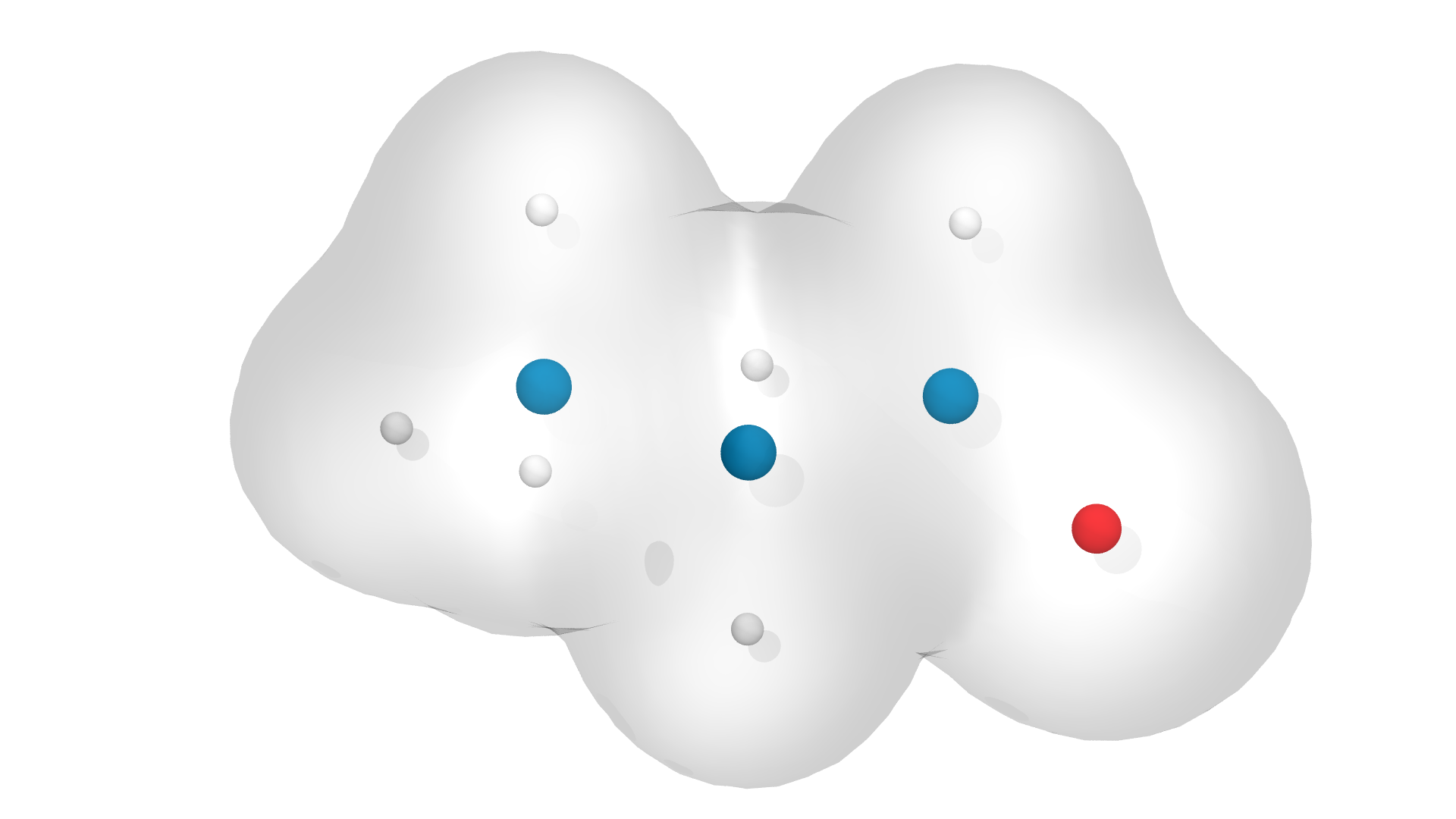
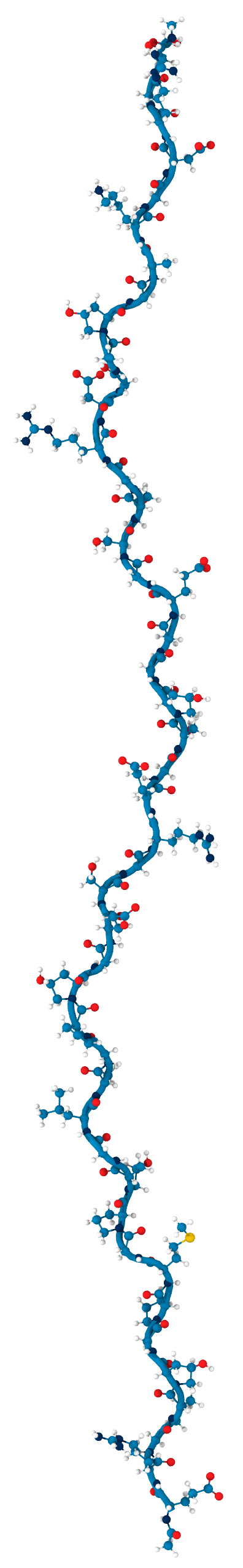
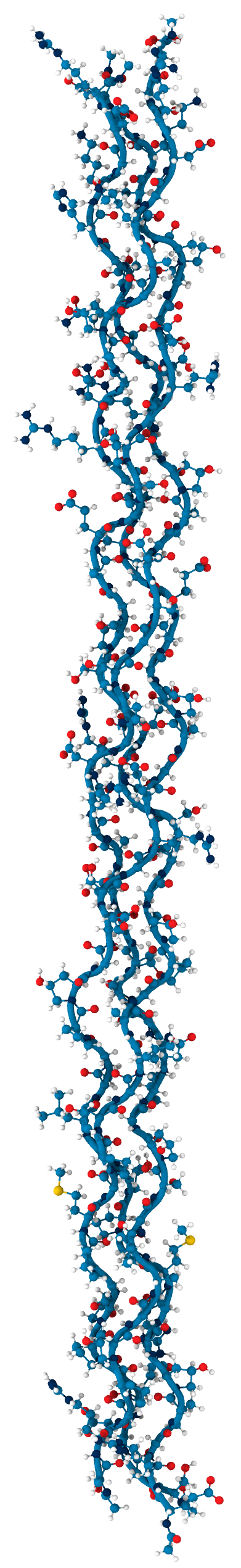
Collagen keeps us together
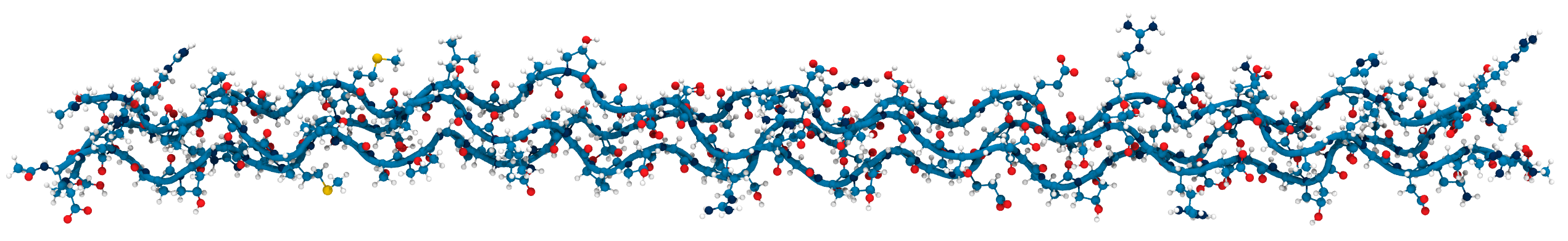

- single peptide
- triple helix
- collagen fibril
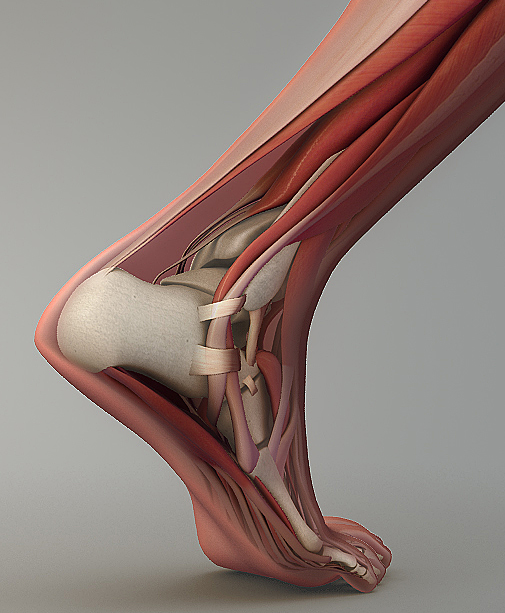
Image sources: foot: Scientfic Animations, https://scientificanimations.com/wiki-images/, SEM: Hughes et al., Figure 5d of [2].
The Gly-X-Y motif allows tight turns and packing
X,Y: proline or hydroxyproline
Prolyl hydroxylase ( 🟠 )
Collagen I is cross-linked
Lysyl hydroxylase
→
( 🟠 )
Lysyl oxidase
→
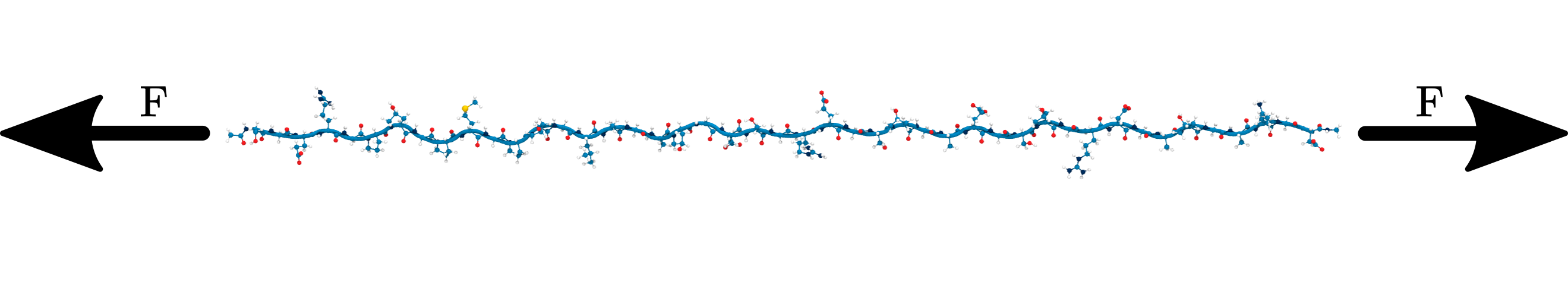
A rather radical discovery: mechanoradicals in collagen

“Mechanoradicals in tensed tendon collagen as a source of oxidative stress”, Zapp et al. (2020) [3]
Goals
for this talk, and by extension, my thesis
Dissecting cleavage mechanisms in tensed collagen fibrils using
reactive molecular dynamics simulations
“Mechanical Activation Drastically Accelerates Amide Bond Hydrolysis,
Matching Enzyme Activity”, Pill et al. (2019) [4]
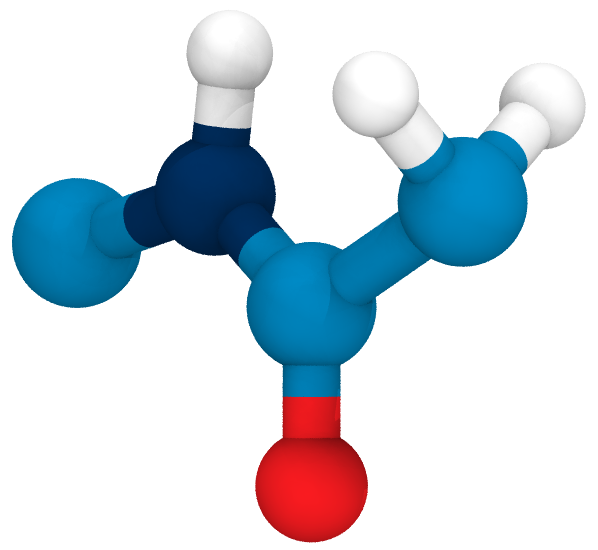
Hydrolysis
vs.
Homolysis
What is the influence of the collagen structure on their competition?
Why do we still see radicals?
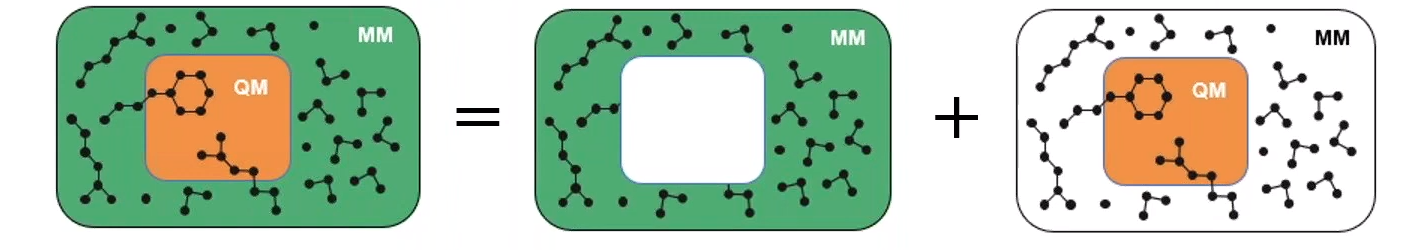
Two complementary approaches
QM/MM simulations of
base-catalyzed hydrolysis
Kinetic Monte Carlo Molecular Dynamics:
KIMMDY
Methods and Theory
Molecular dynamics / Molecular Mechanics
How do we make the atoms move?
\[ \begin{array}{rcl} F &=& ma \\ -\frac{dV}{dr} &=& m\frac{d^2r}{dt^2}, \end{array} \tag{1}\]
\(V\): potential
\(r\): position
\(F\): force
\(a\): acceleration
\(t\): time
\(m\): mass
Force Fields and Topologies
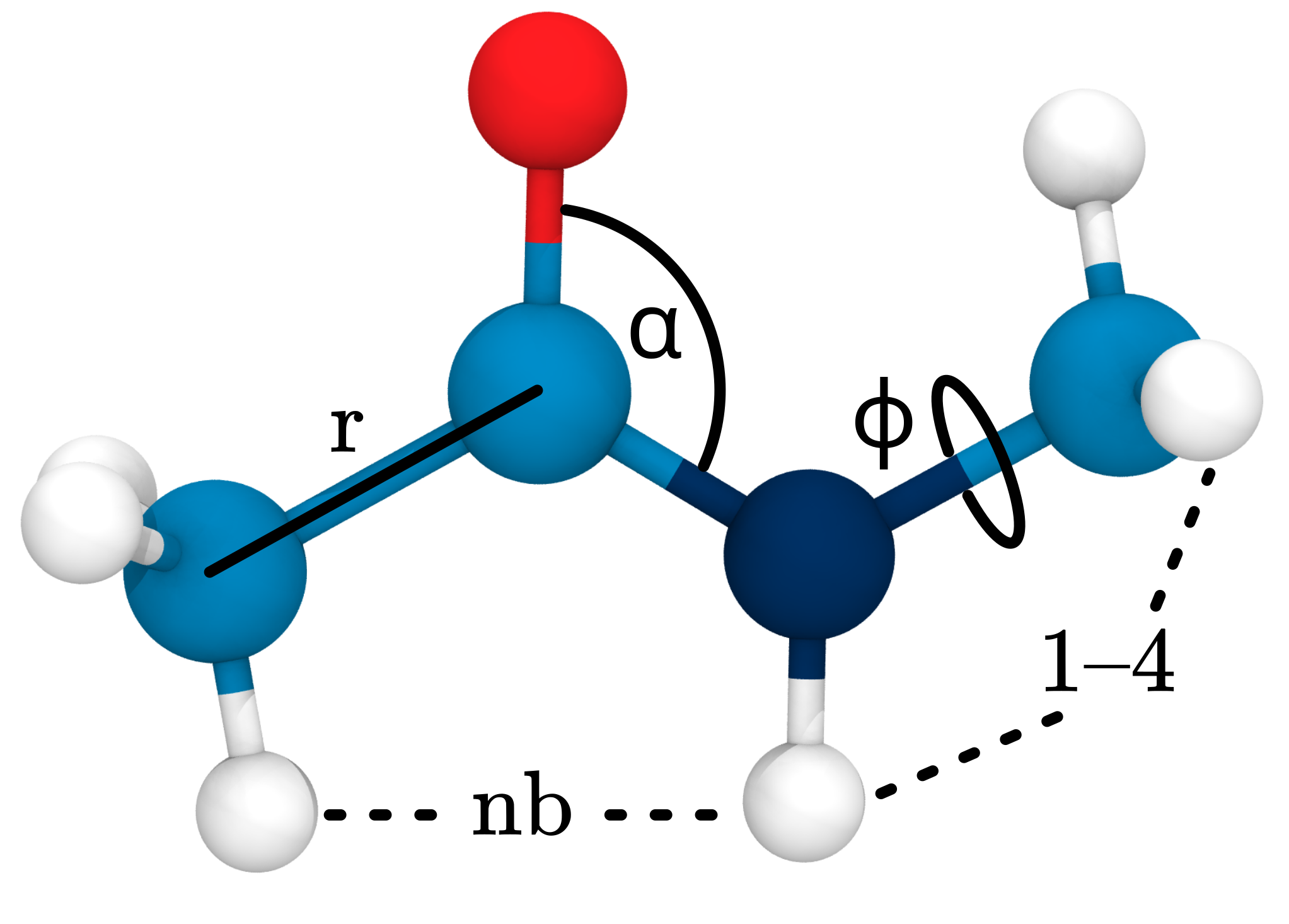
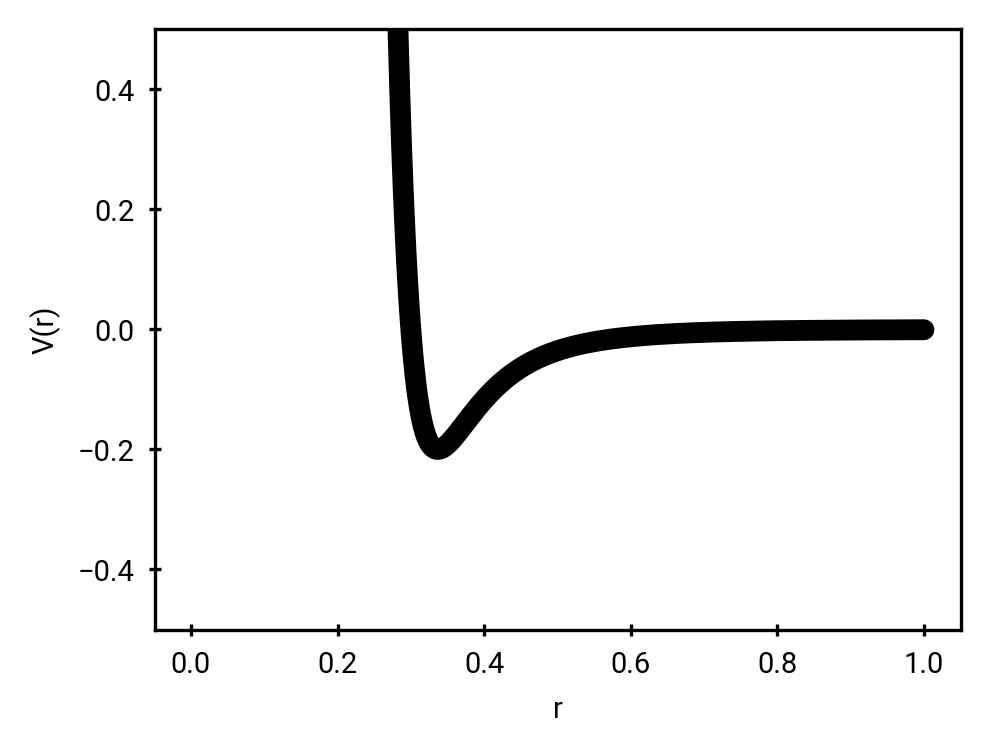
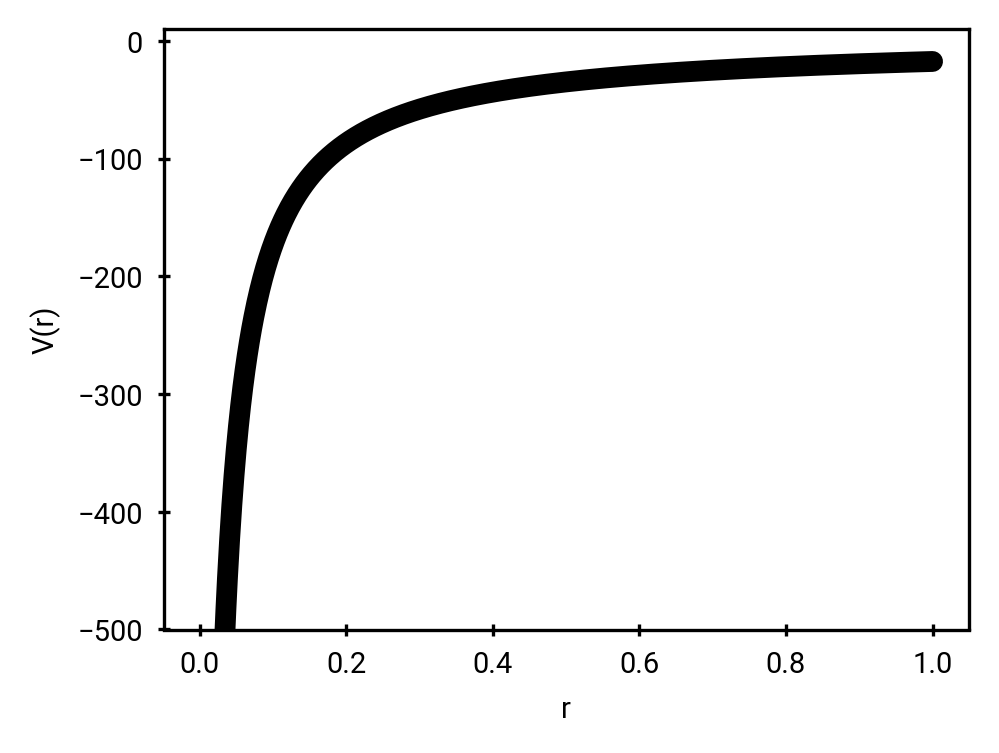
Connectivity and parameters pre-defined!
So how do we do chemistry?
Quantum mechanics/molecular mechanics


DFT: Density Functional Theory
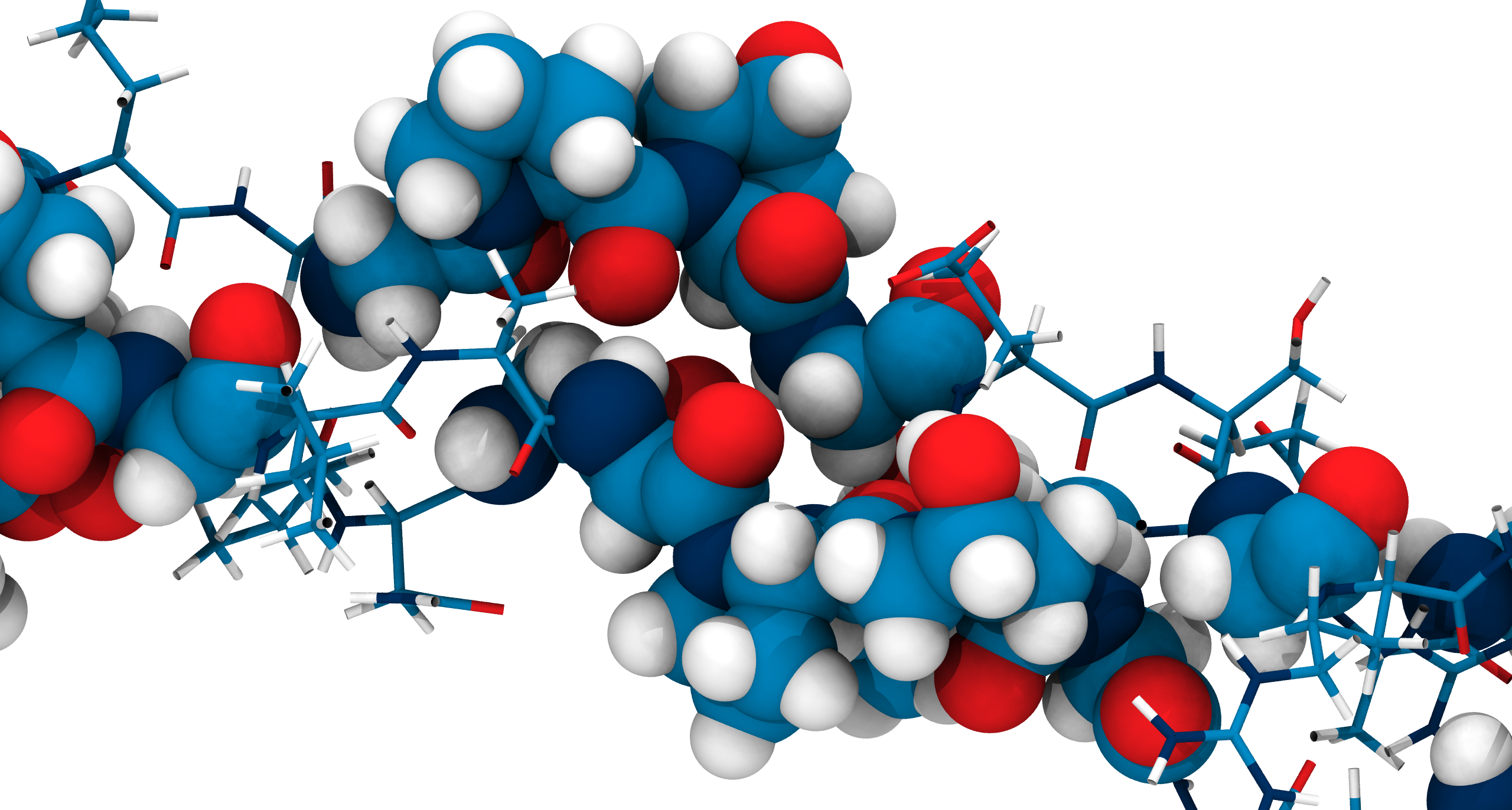
Collagen Hydrolysis by Quantum Mechanics/Molecular Mechanics
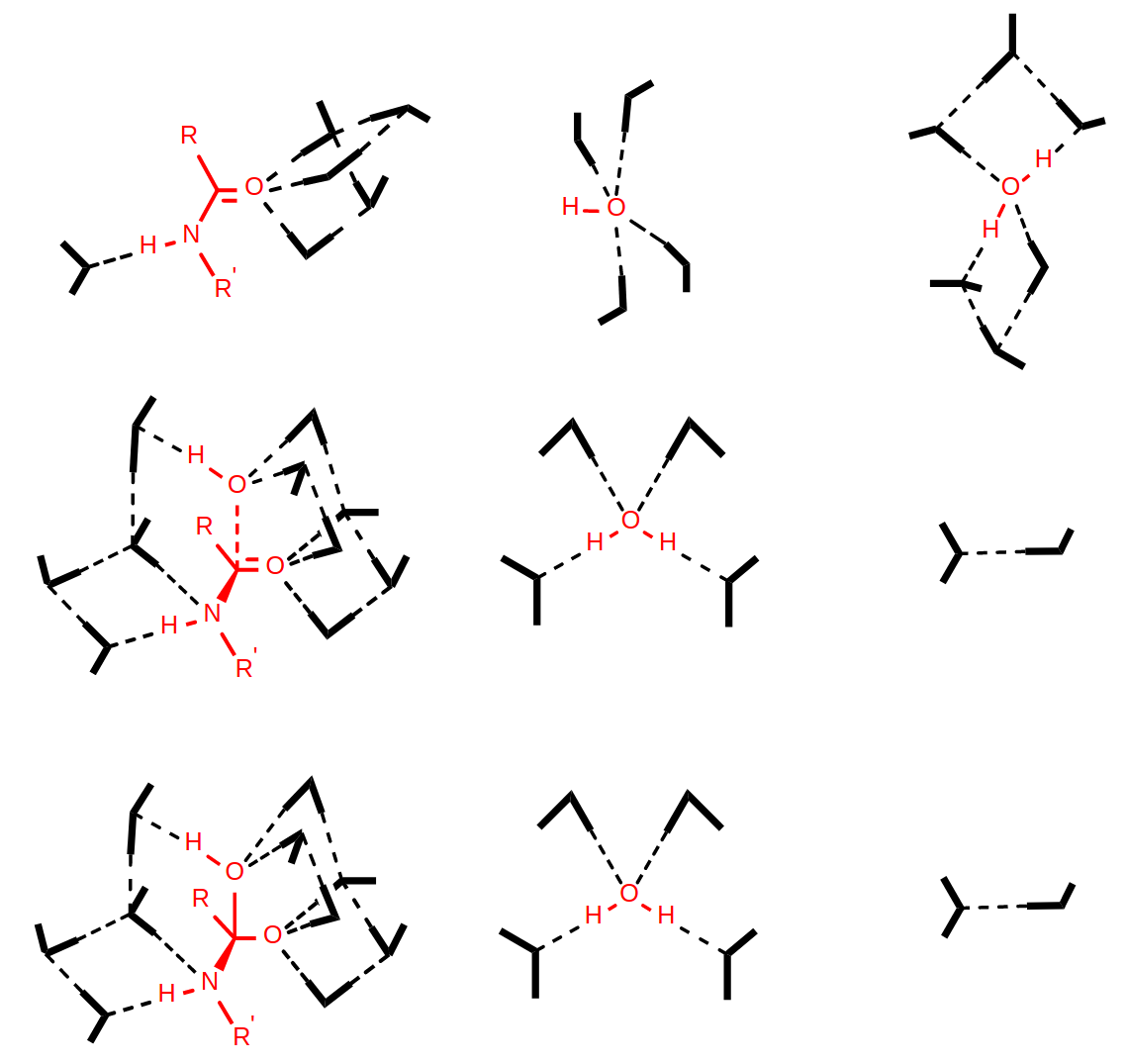
Base-catalyzed hydrolysis
TS: transition state
TI: Tetrahedral Intermediate
ZI: Zwitterionic Intermediate
Previous work on base-catalyzed hydrolysis
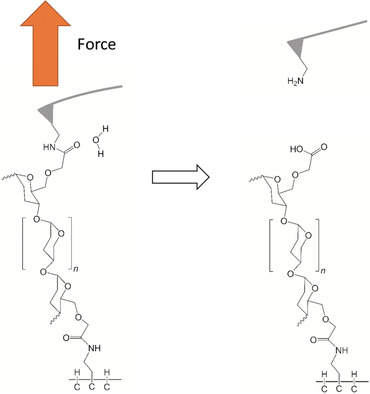
System Setup
[]: input parameters.QM region selection and setup
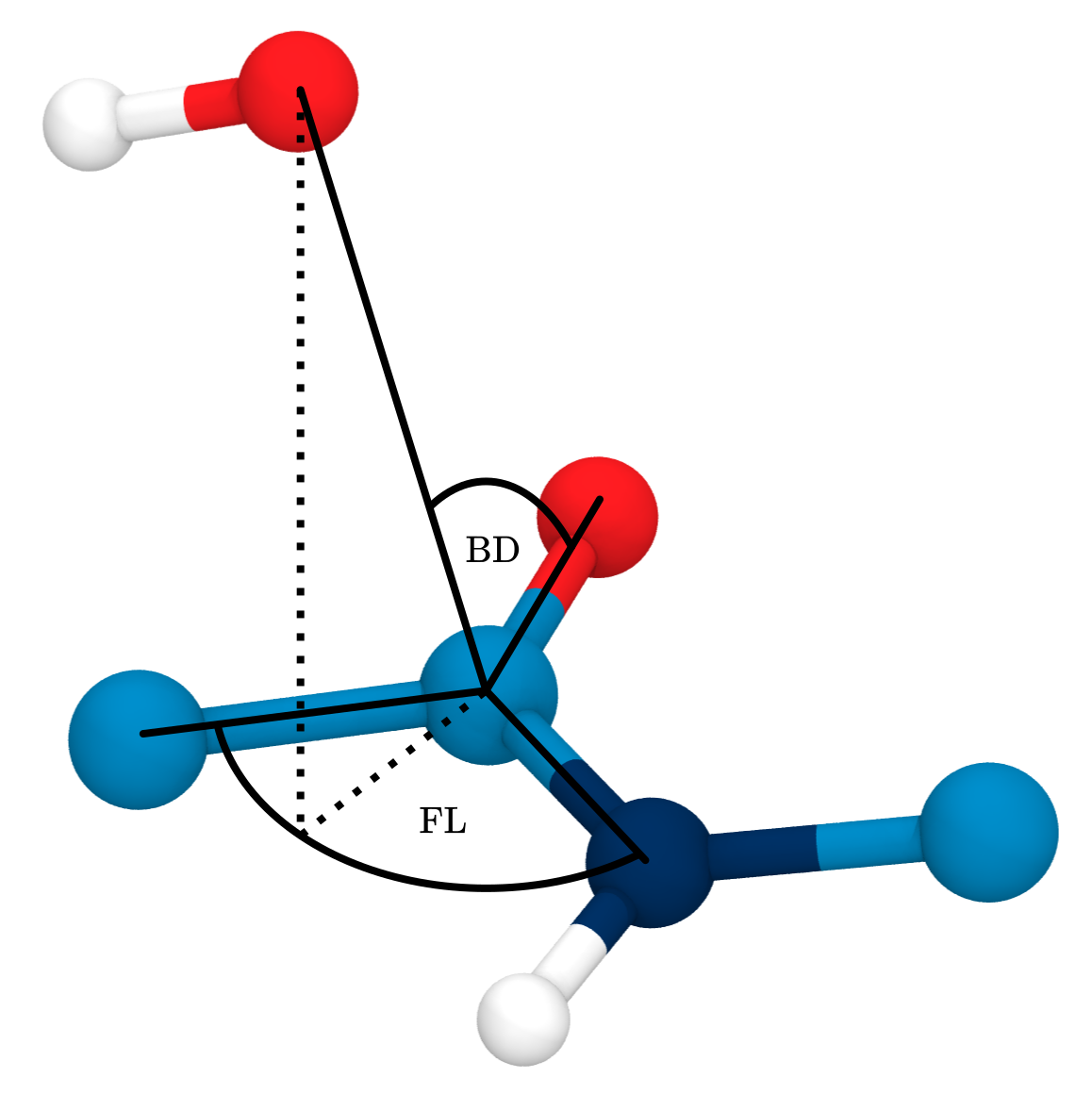
\(BD\): Bürgi-Dunitz angle, 107°
\(FL\): Flippin-Lodge angle, 0°
Umbrella sampling the reaction coordinate

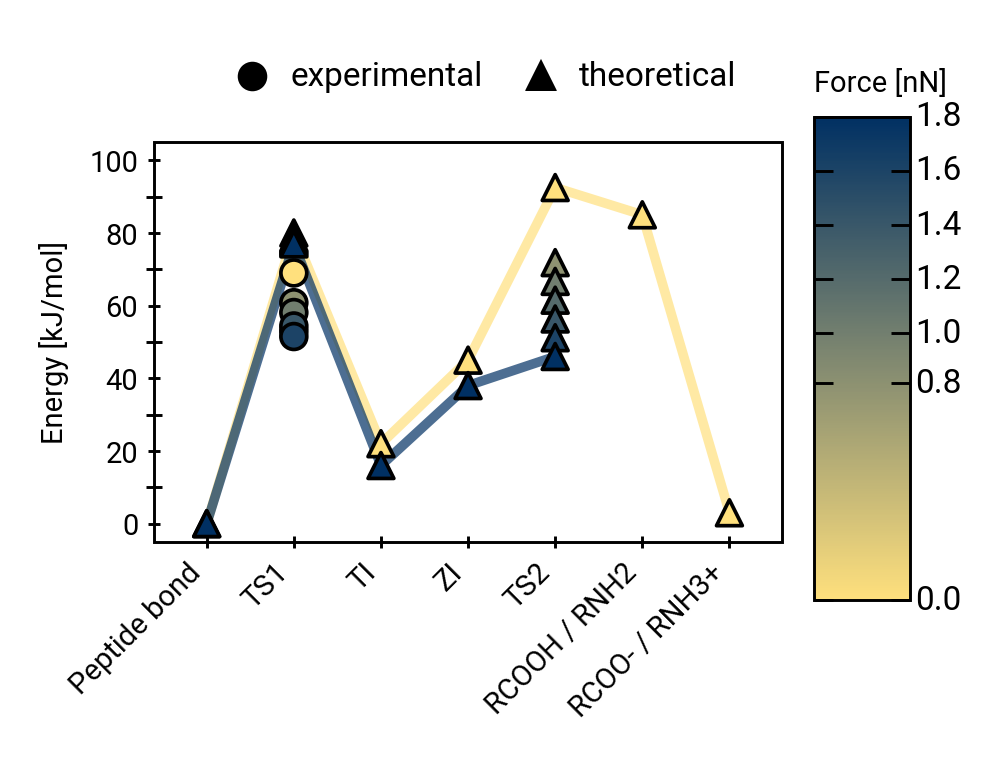
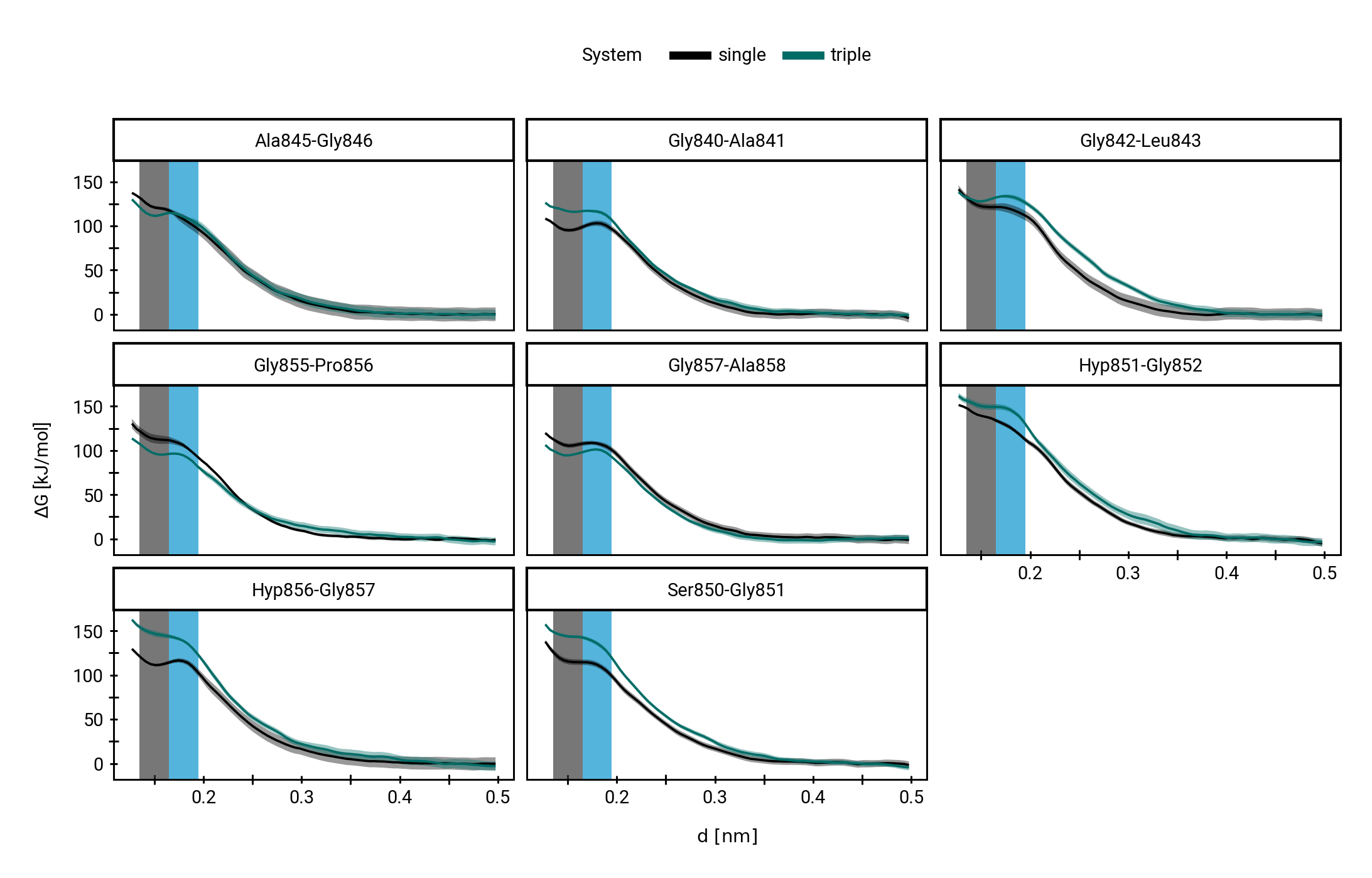
QM/MM simulations reveal mechanistic details of the tetrahedral intermediate formation
QM/MM simulations reveal mechanistic details of the tetrahedral intermediate formation

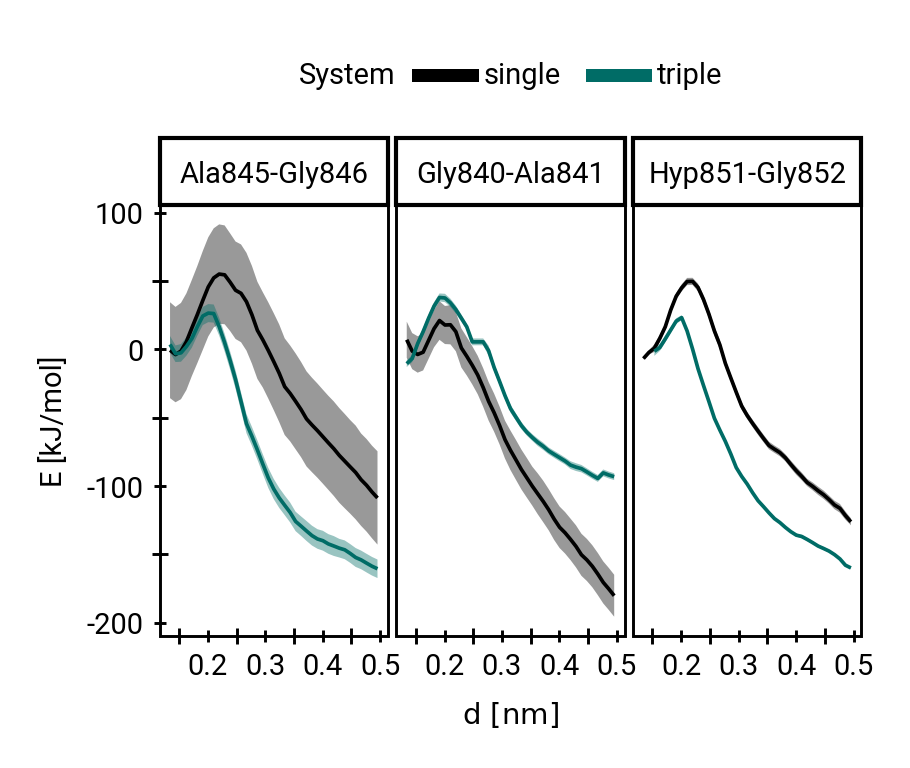
The triple helix marginally impedes hydroxide attack thermodynamically
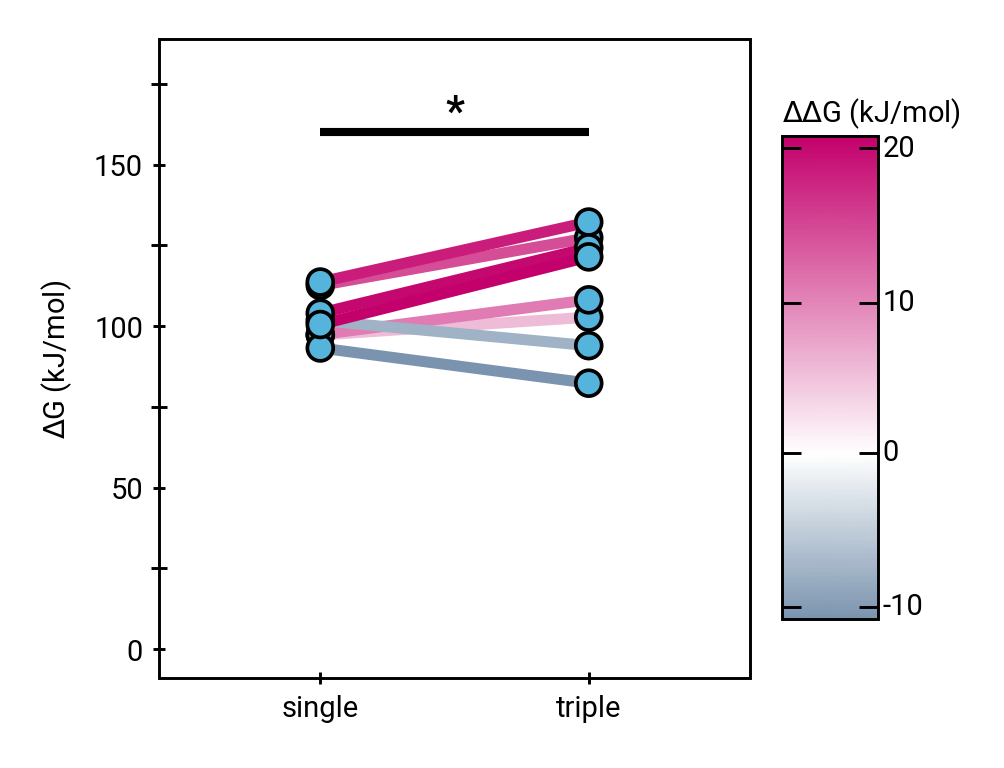
\(\Delta\Delta G\) 9.02 ±4.41 kJ/mol (±SEM)
Rate reduction due to triple helix:
30 to 40 times.
WHAM: Weighted Histogram Analysis Method [6]

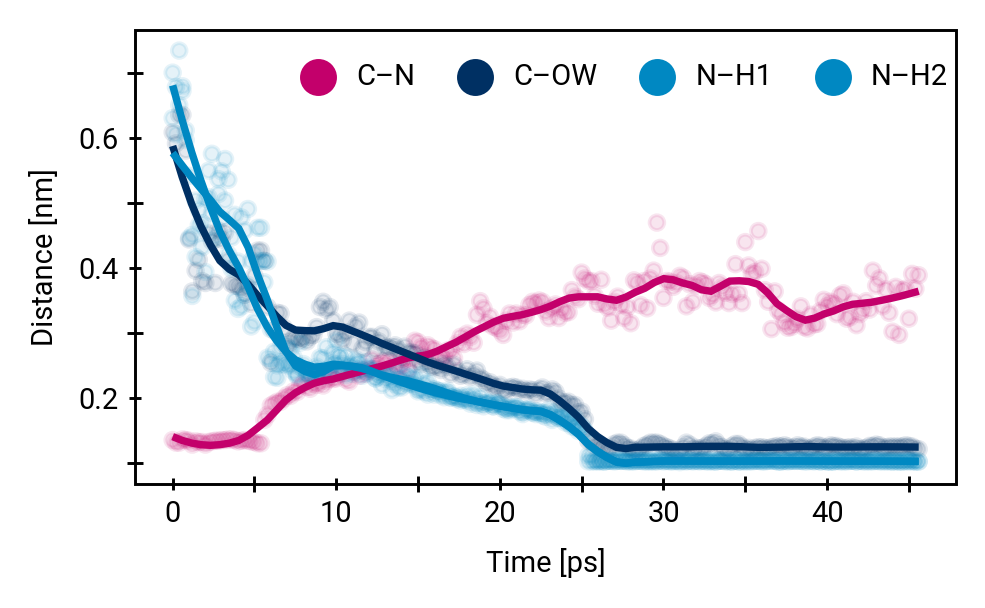
Proton mobility in explicit QM solvent molecules allows breaking of the peptide bond
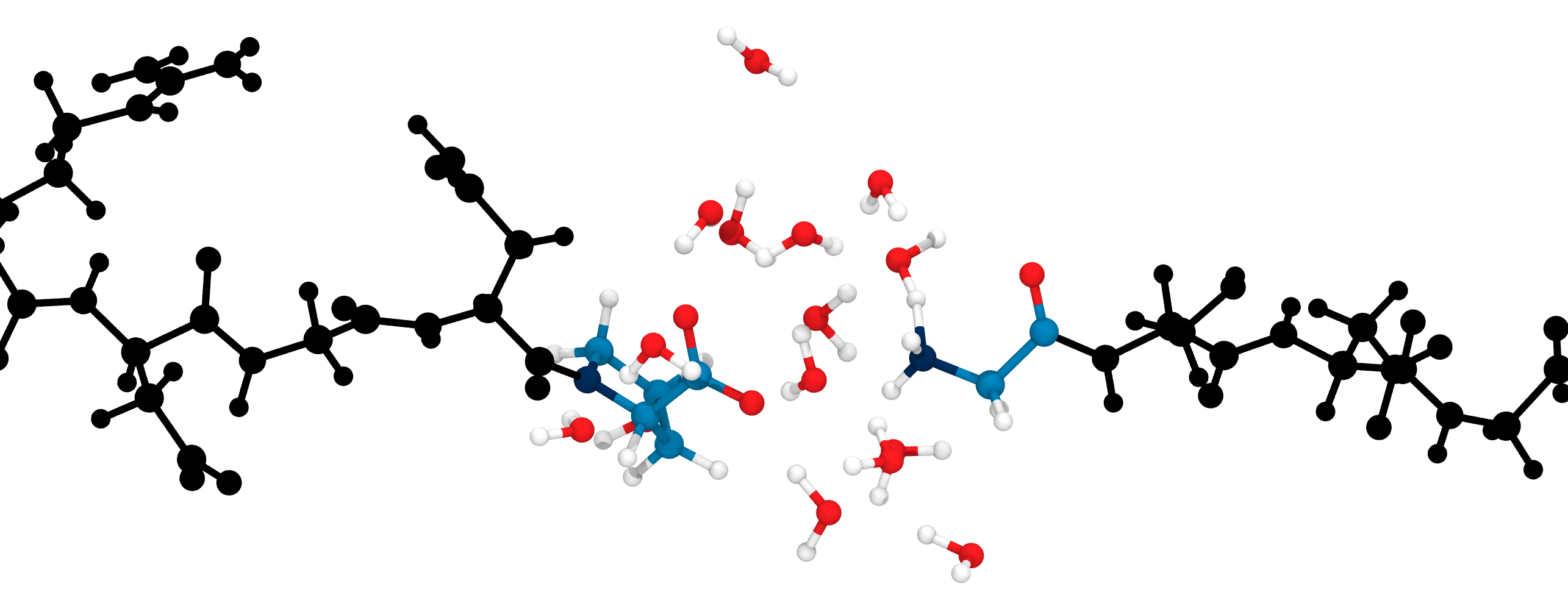
Discussion of QM/MM results
- Currently in peer review at The Journal of Physical Chemistry B
- Survivorship bias

- Effect size

\(\Delta\Delta G\) 9.02 ±4.41 kJ/mol (±SEM)
- Computational cost!
Image source Airplane: Martin Grandjean (vector), McGeddon (picture), US Air Force (hit plot concept), CC BY-SA 4.0, https://commons.wikimedia.org/w/index.php?curid=102017718
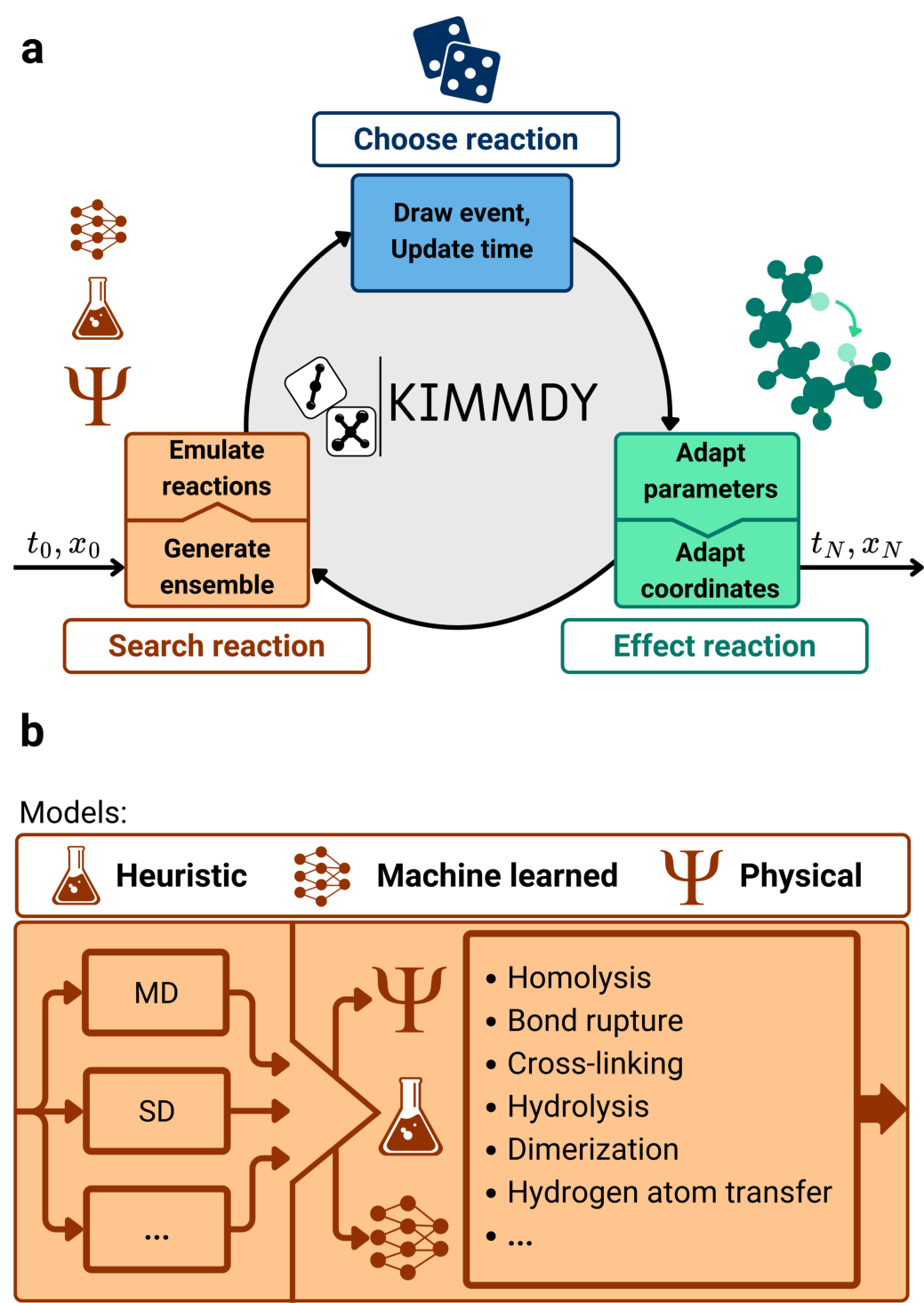
Kinetic Monte Carlo Molecular Dynamics: KIMMDY
Kinetic Monte Carlo Molecular Dynamics: KIMMDY

with Eric Hartmann and Kai Riedmiller,
inspired by Benedikt Rennekamp [7]
graeter-group.github.io/kimmdy/
Read now on bioRxiv:
KIMMDY: A biomolecular reaction emulator [8],
or soon in Nature Communications.

Schema made by Denis Kieswetter for [8]
doi:10.1101/2025.07.02.662624
KIMMDY is easy to extend to new chemistries

KIMMDY takes care of the complicated topology modifications.
Parametrization interface to obtain parameters after a reaction
Graph Attentional Protein Parametrization by Leif Seute [9]
also a good excuse to show the mol* plugin I wrote for our documentation
Slow-growth to execute even complex reactions like hydrolysis
Does not have to follow the chemically
accurate reaction coordinate
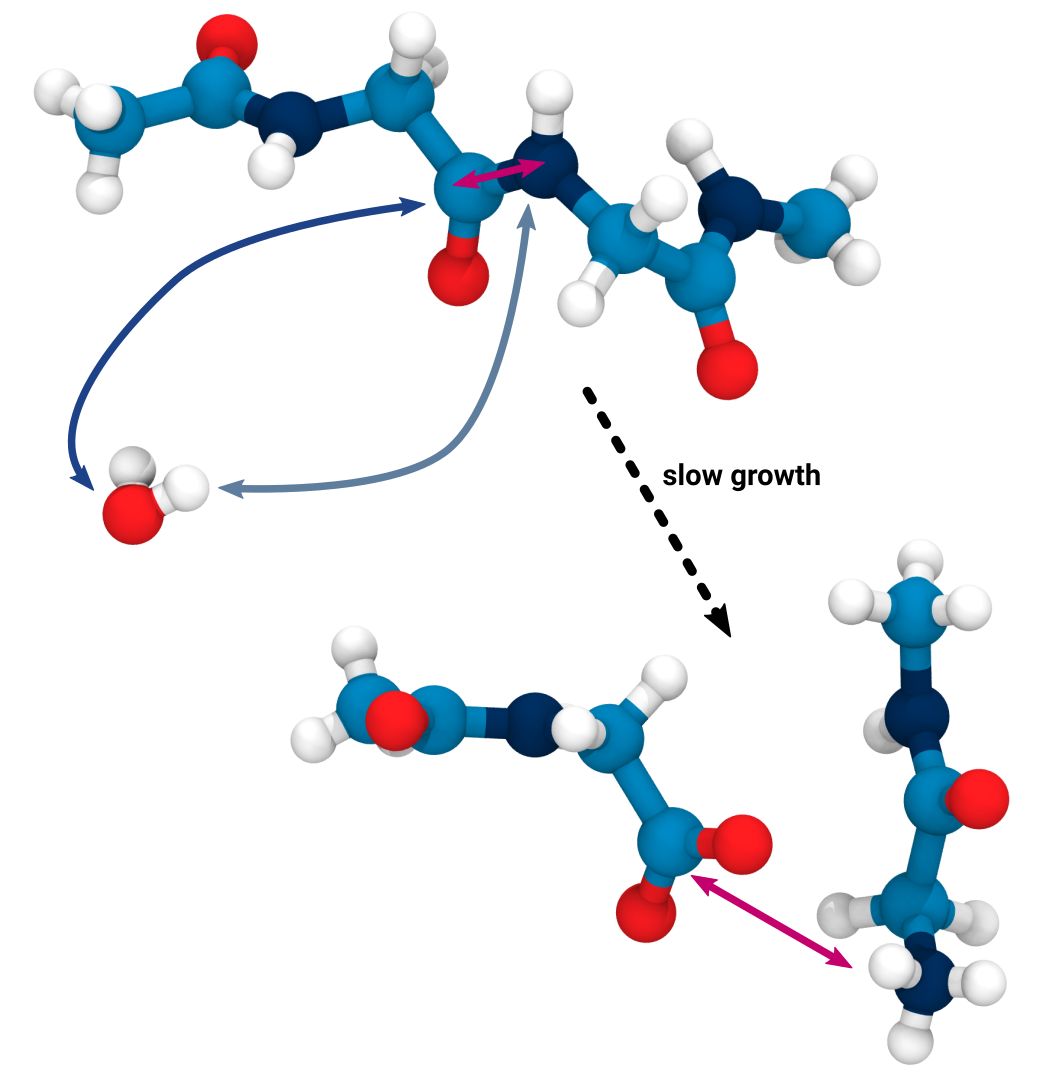
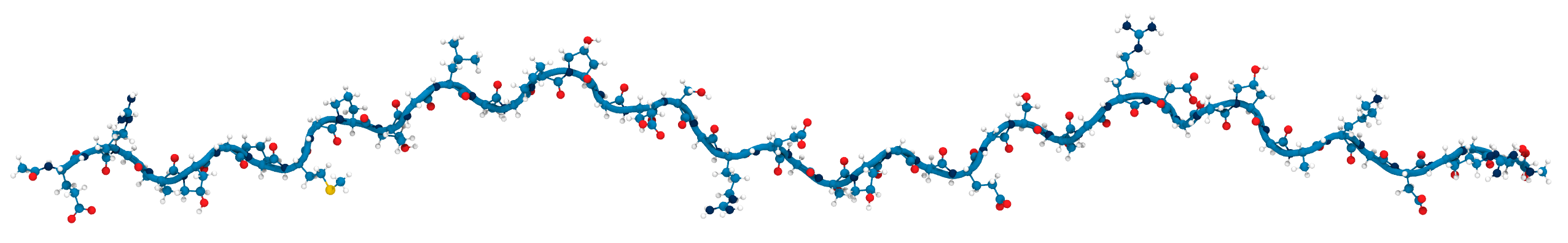
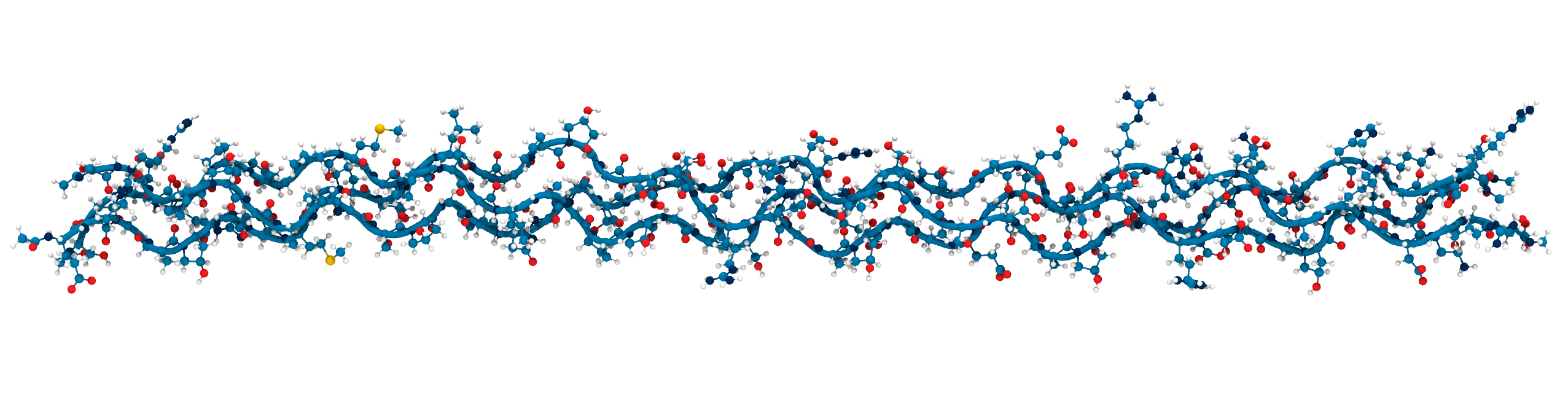
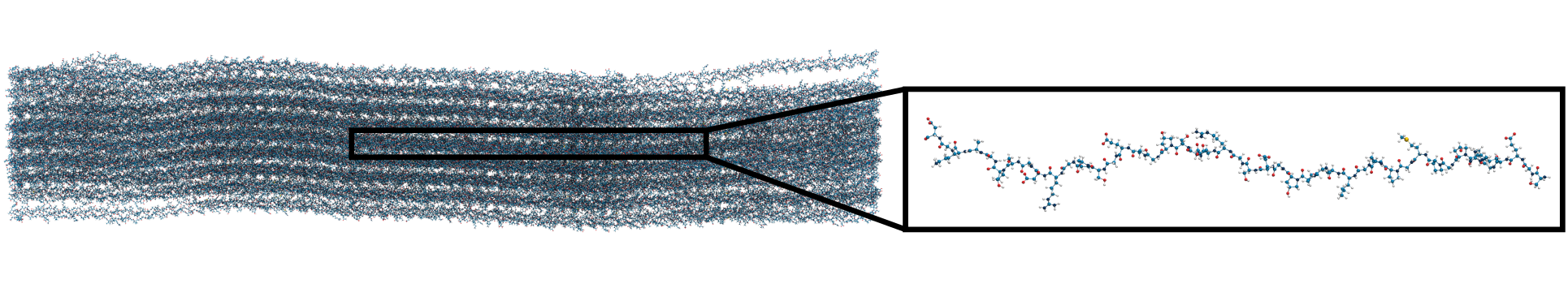
Applying KIMMDY to collagen
hydrolysis and homolysis
Applying KIMMDY to collagen hydrolysis and homolysis

Applying KIMMDY to collagen hydrolysis and homolysis
Arrhenius equation
\[ k = A\,\mathrm{e}^{\left(\frac{-\mathbf{E_\mathrm{a}}}{RT}\right)} \]
\(A\): pre-exponential factor (attempt frequency)
\(R\): gas constant
\(T\): temperature
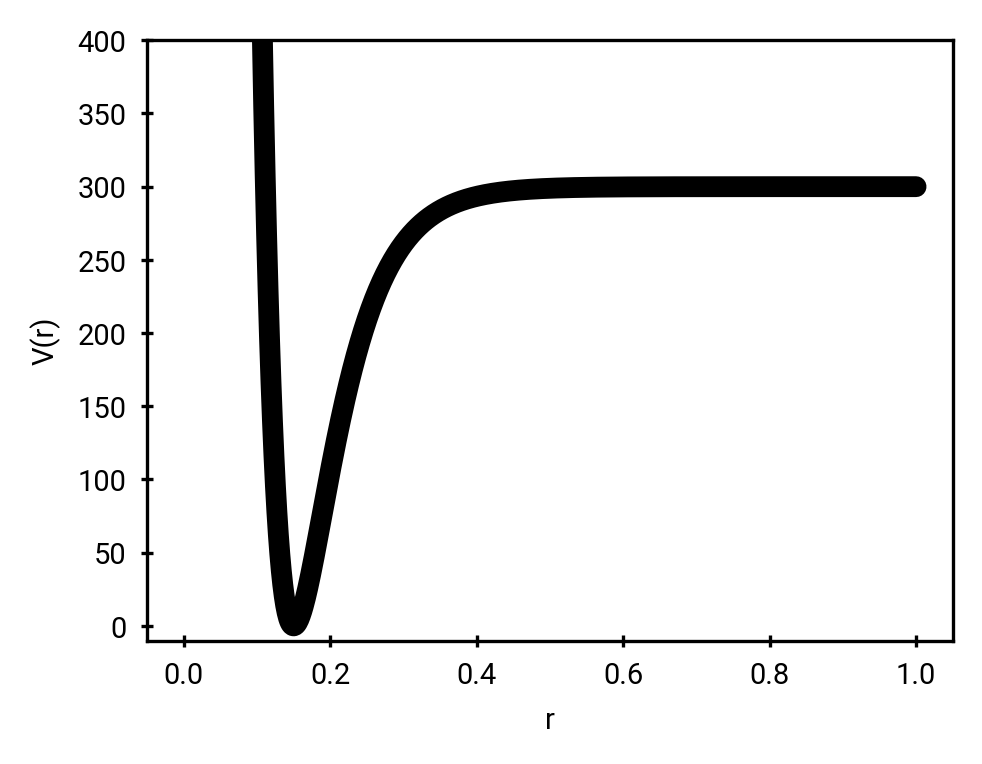
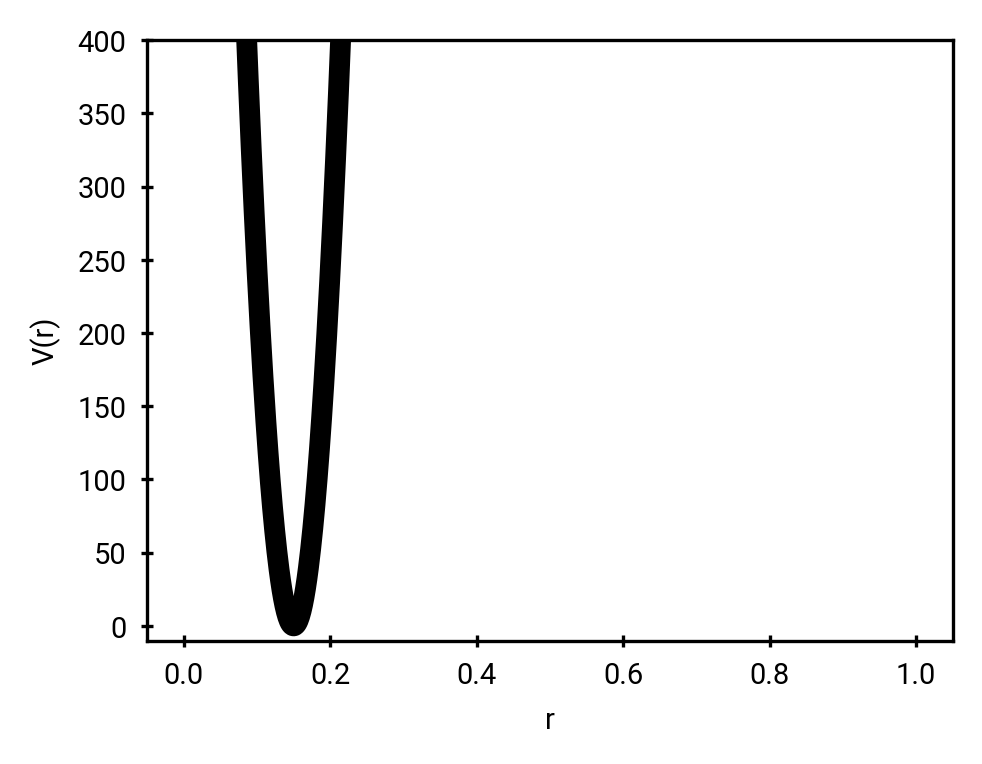
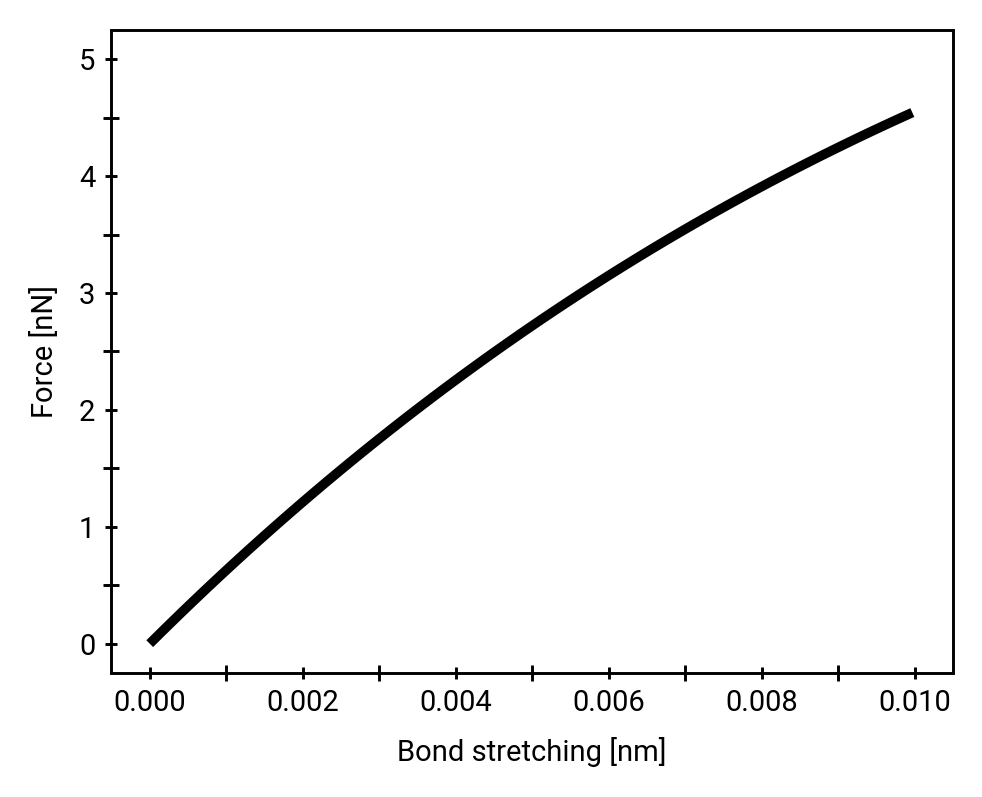
Morse potential

Hydrolysis rate incorporates SASA
SASA

Hydrolysis rate incorporates SASA
SASA = Solvent Accessible Surface Area



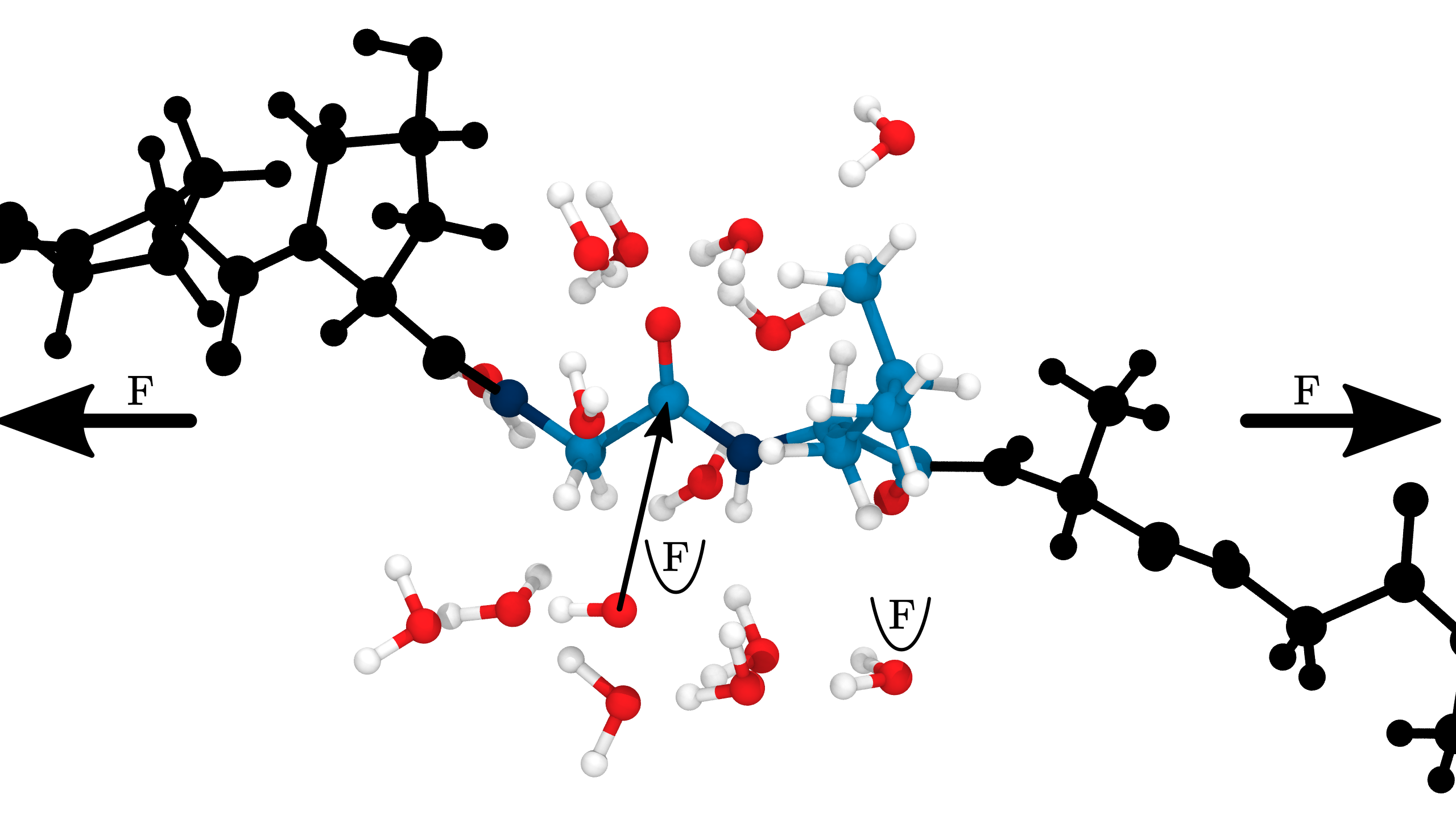
Hydrolysis rate incorporates pH, SASA and force


\[ \mathrm{Rate}(bond, t) = \ \frac{\mathrm{SASA}(bond, t)}{\mathrm{SASA}_{max}} \cdot \ \frac{c_{OH^-}}{c_{OH^-_{\mathrm{exp}}}} \ \cdot \mathrm{rate}_{\mathrm{exp}}(F_{bond,t}) \tag{2}\]

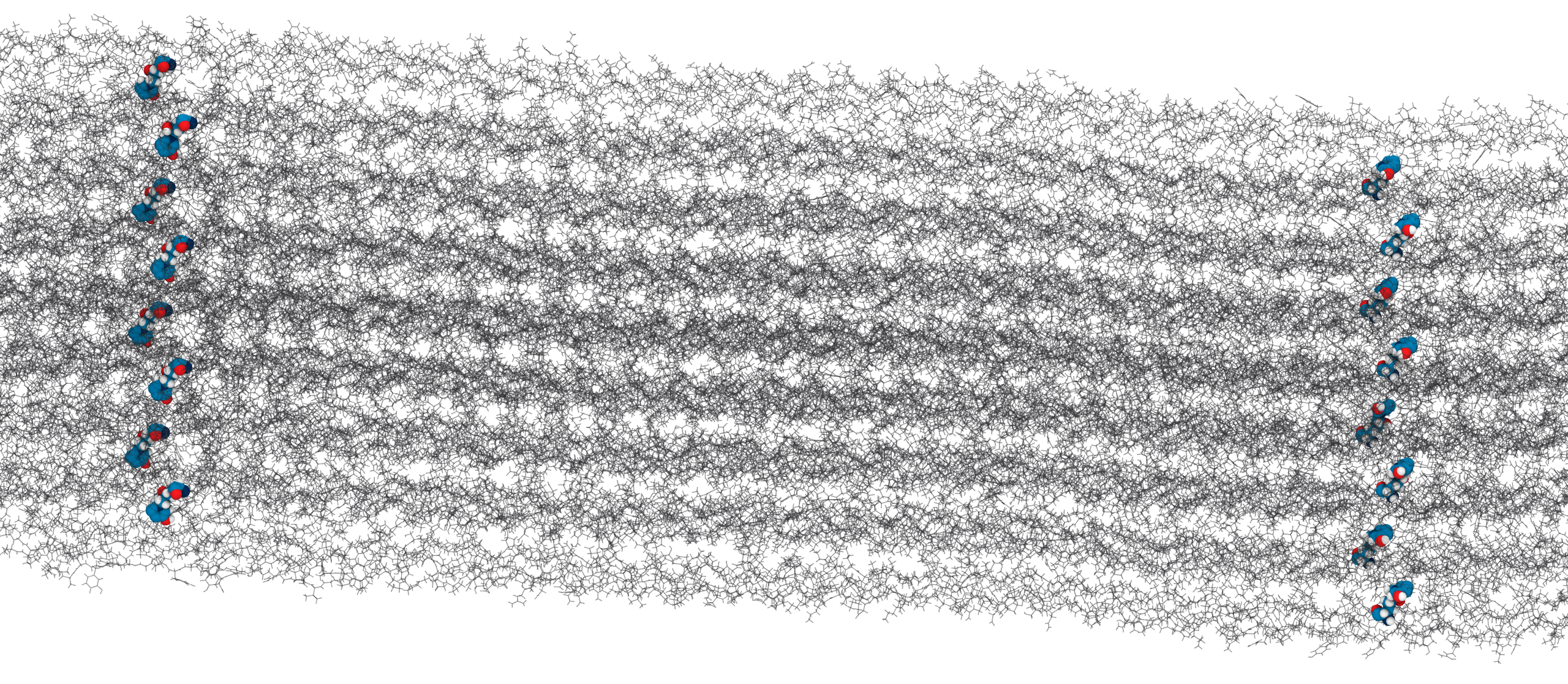
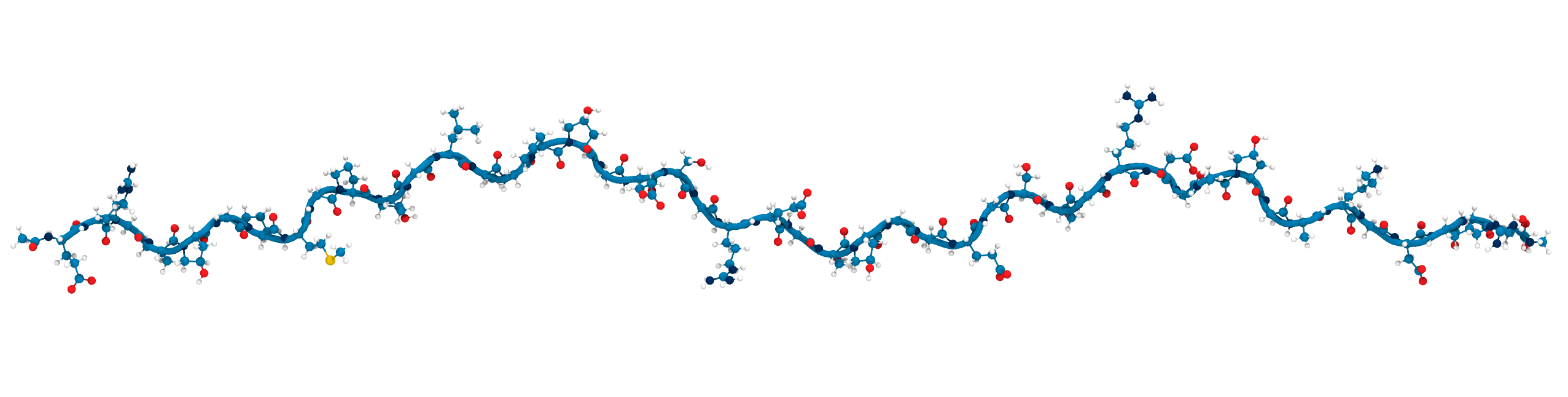
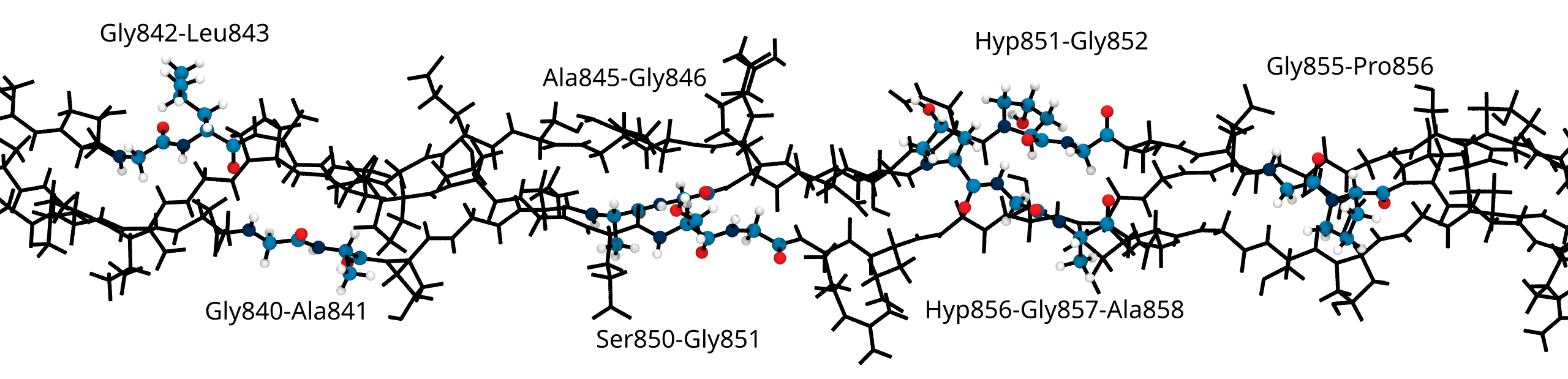
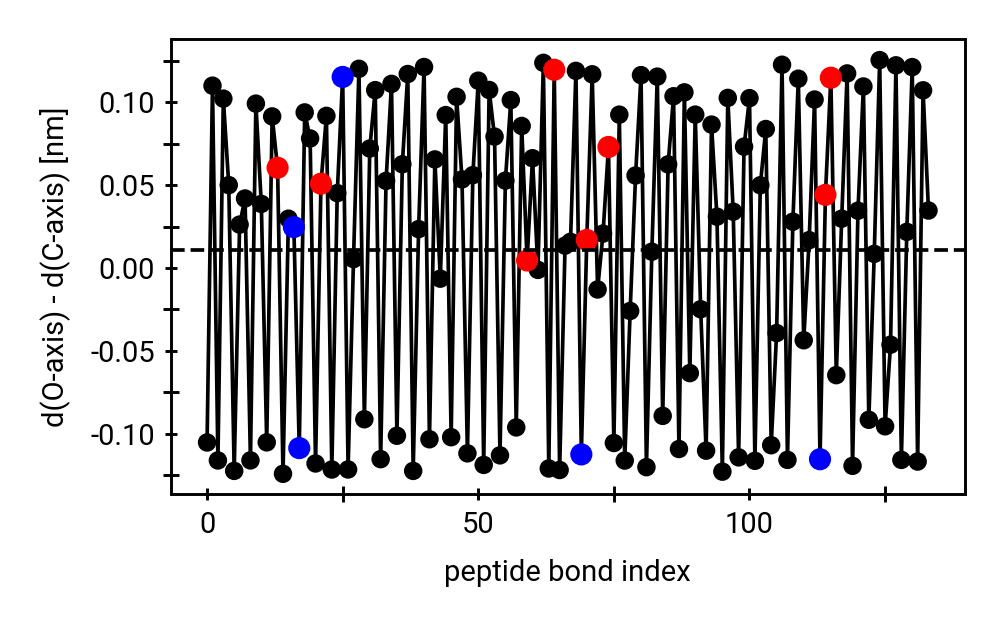
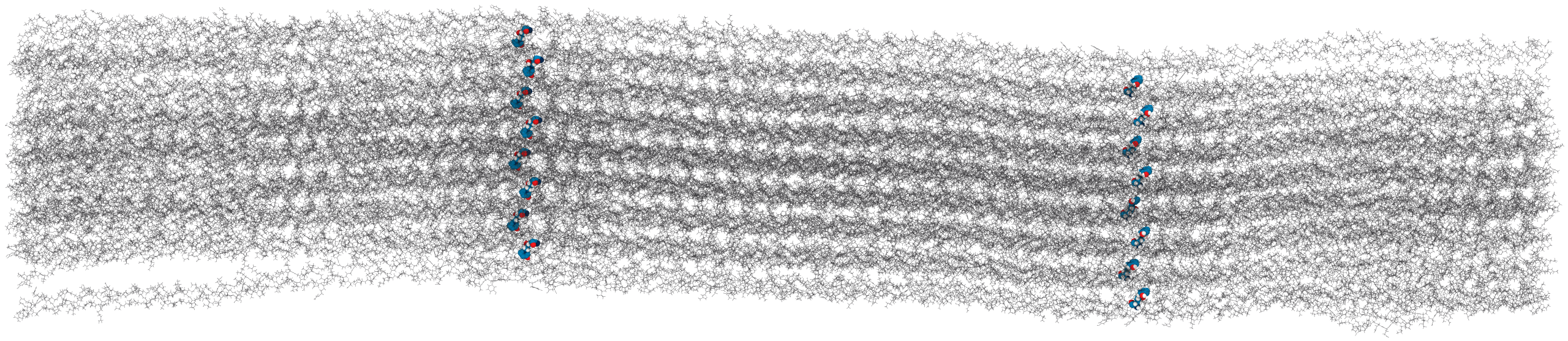
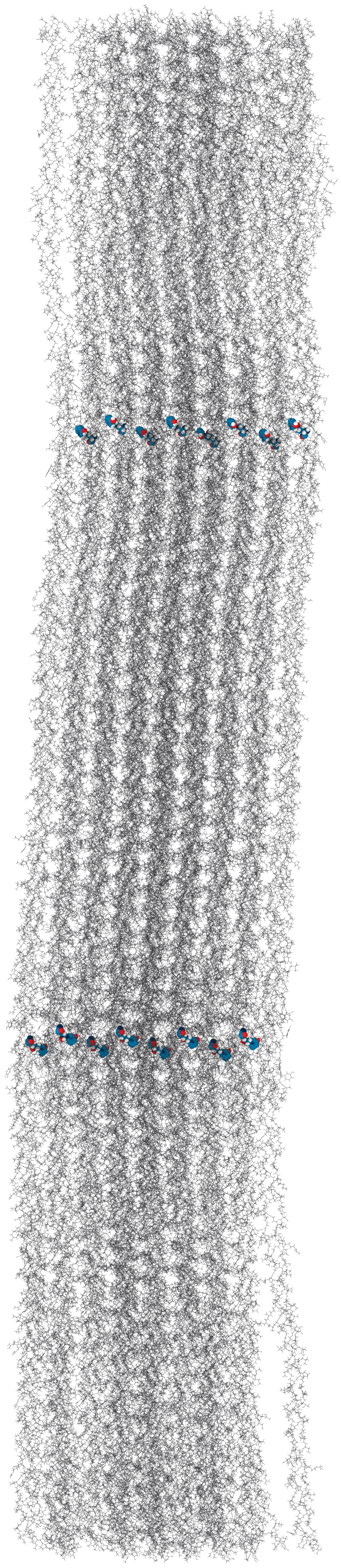
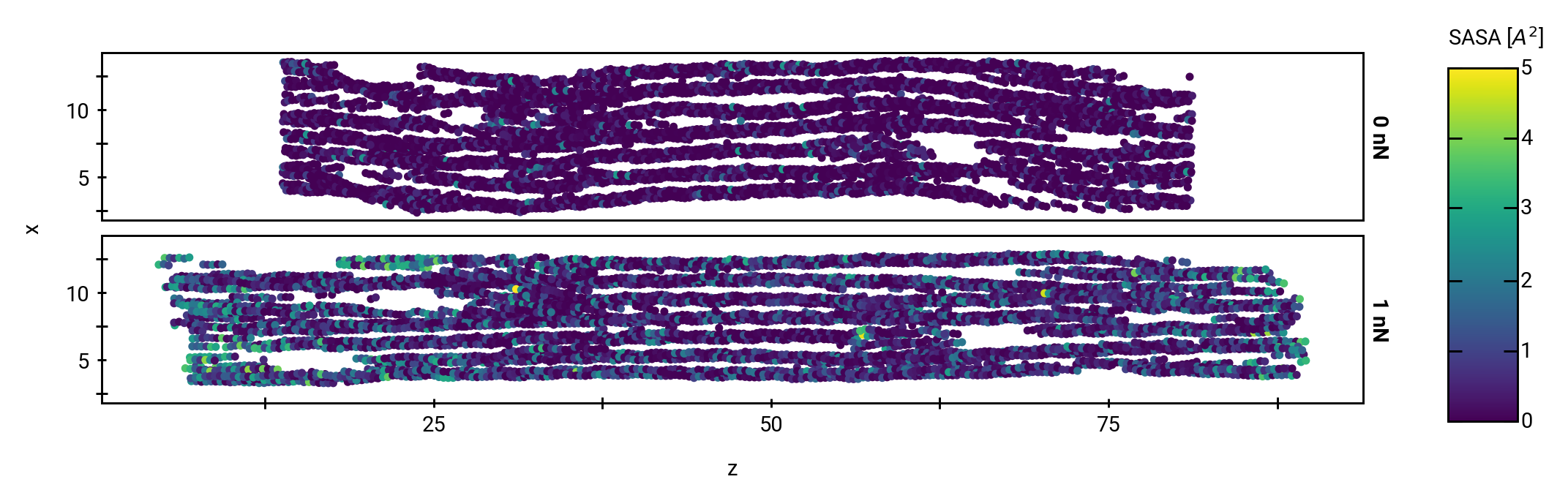
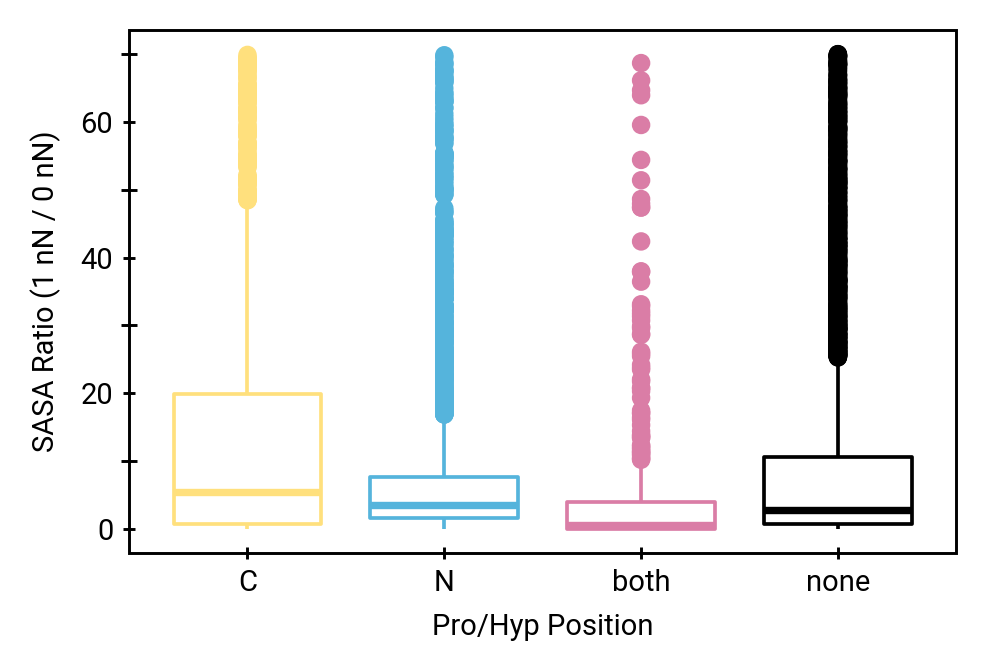
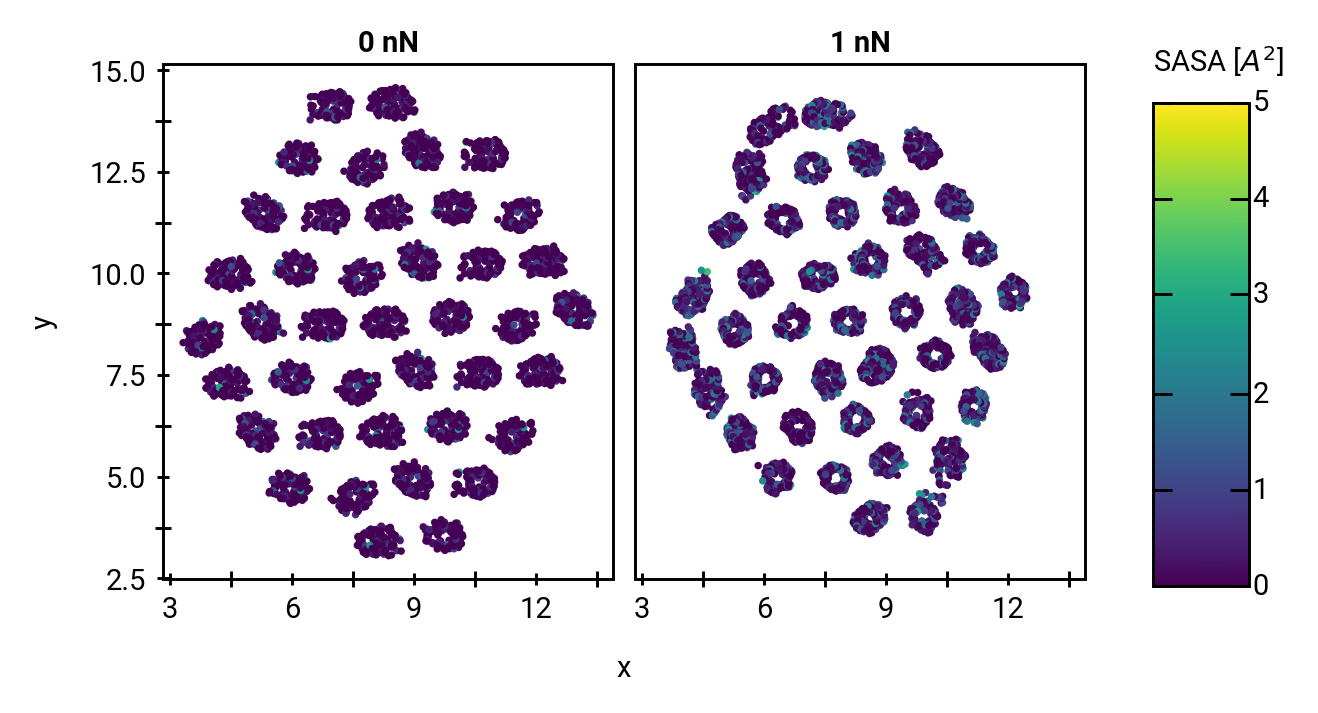
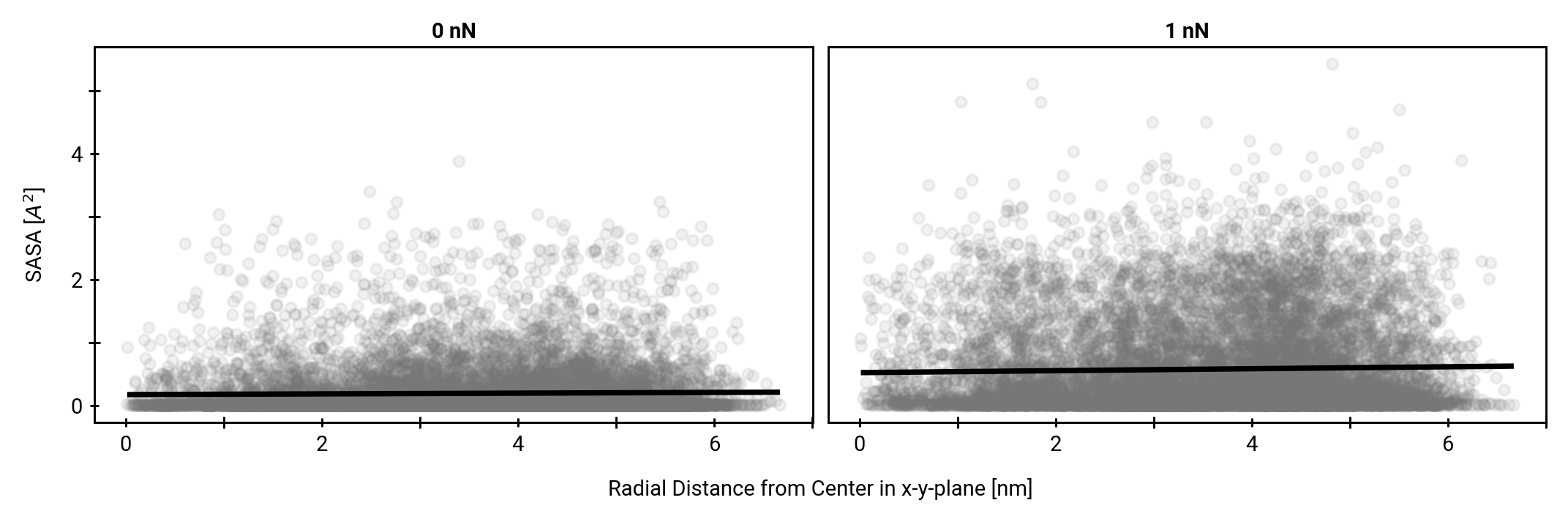
Solvent accessibility in the fibril is heterogeneous
peptide bonds near the center are still accessible
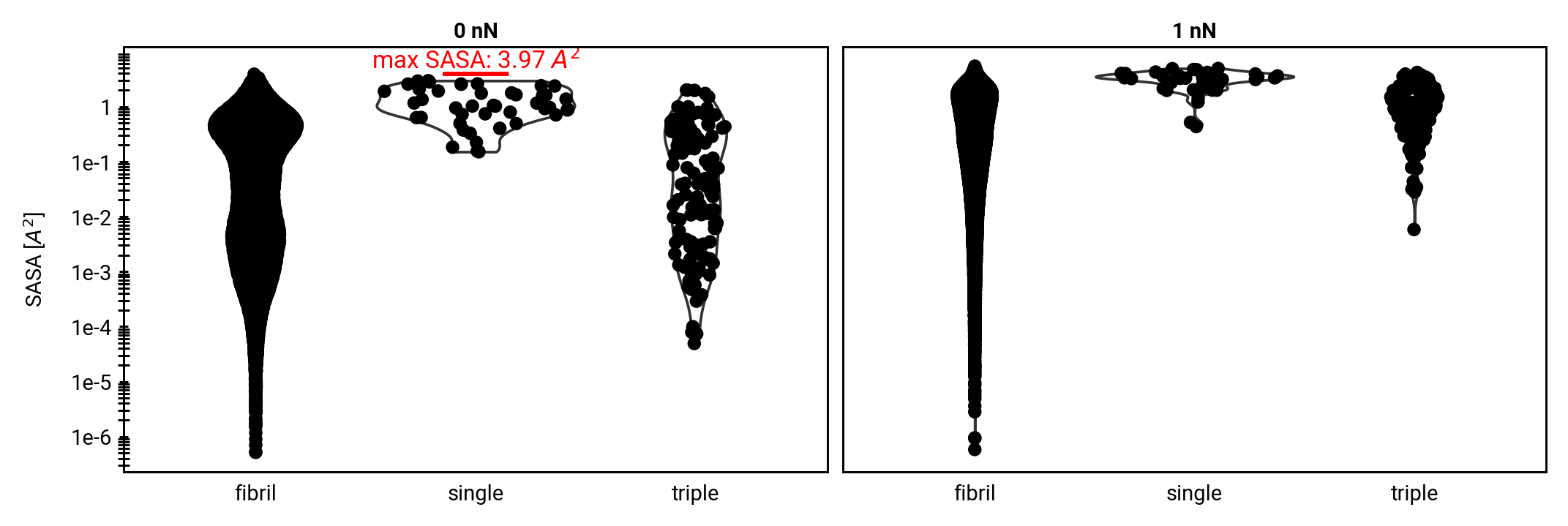



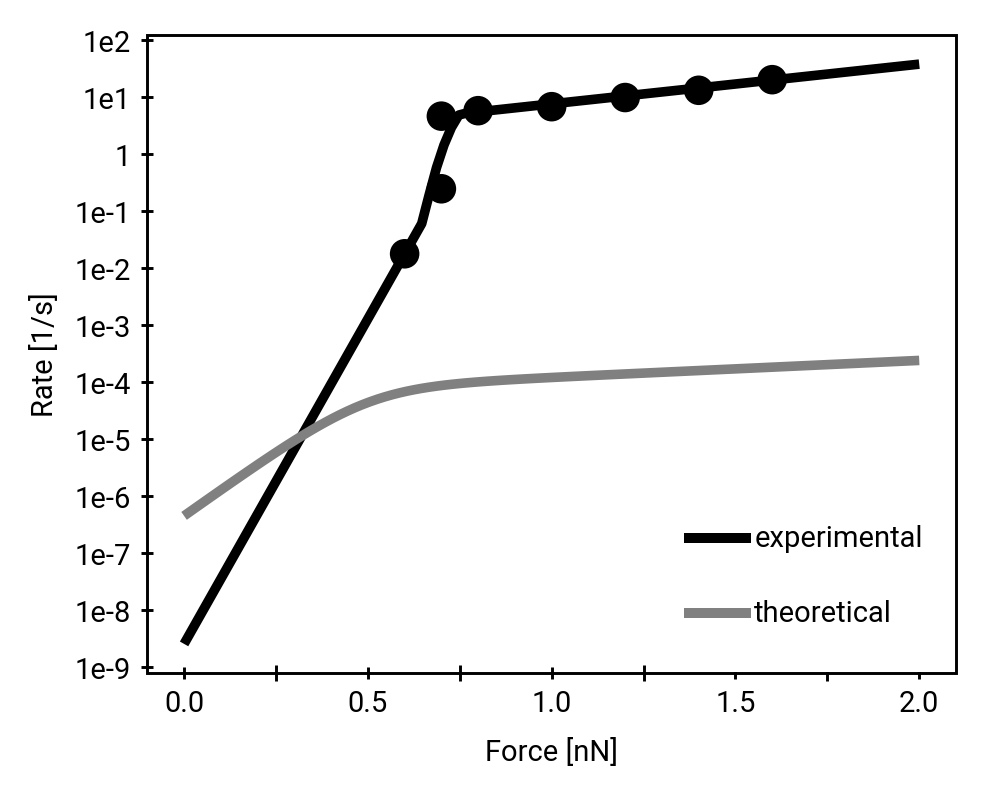
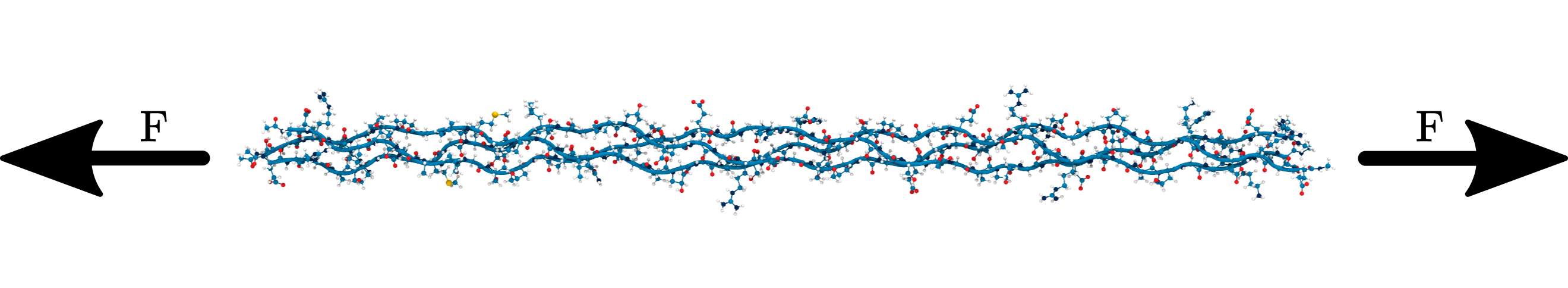
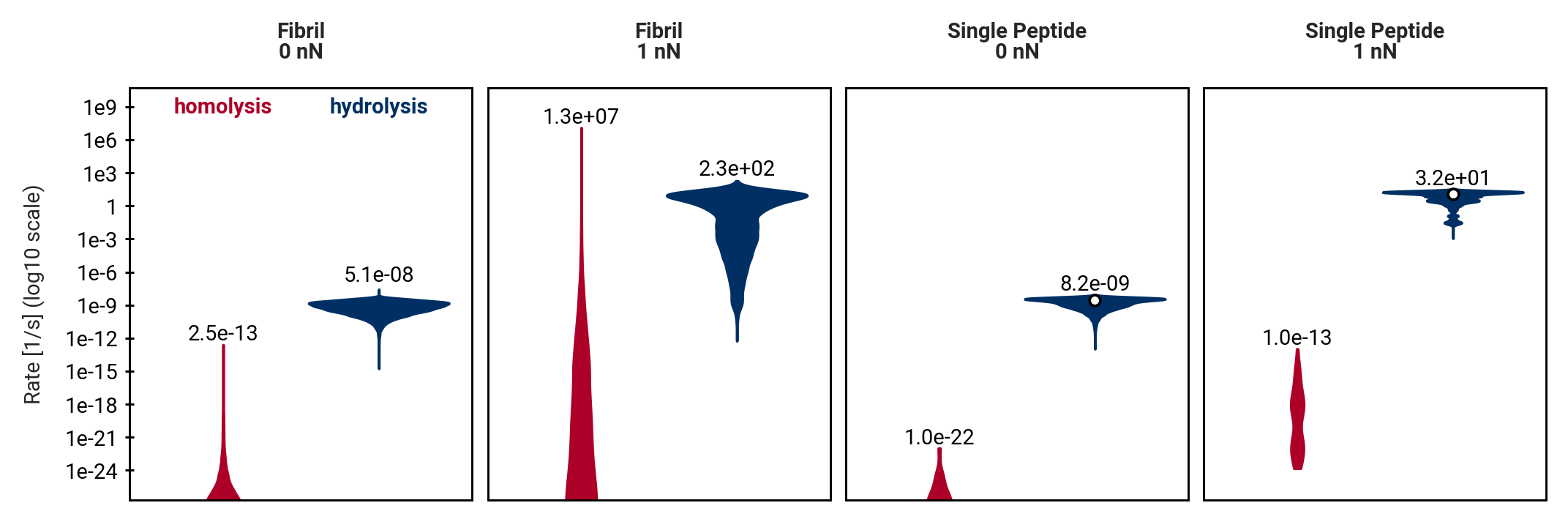
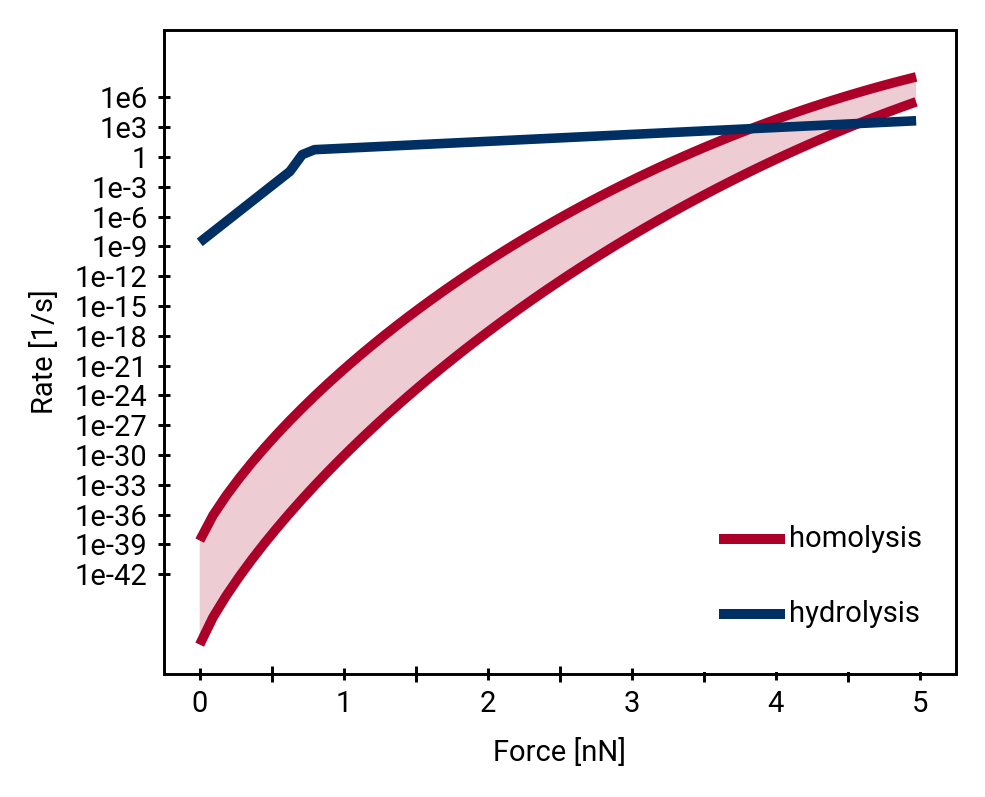
Homolysis becomes competitive with hydrolysis by force concentration
Discussion of KIMMDY results
- pH just a factor, could be e.g. constant pH simulations [12]
- SASA influence likely to be underestimated
- Need a link to real tissue experiments

Concluding Thoughts
Concluding Thoughts
KIMMDY is a versatile tool for reactive MD simulations:
graeter-group.github.io/kimmdy/

Collagen triple helix impedes hydrolysis


Homolysis becomes competitive to hydrolysis due to force concentration in cross-linked collagen fibrils

Thank You!
Thank You!
Questions?


This work was supported by the Klaus Tschira Foundation and has received funding from the European Research Council (ERC).
References
Appendix
Publications not included in this presentation
- Buhr et al., Intrinsically disordered region of talin’s FERM domain functions as an initial PIP2 recognition site, published at Biophysical Journal (2022), 10.1016/j.bpj.2023.02.020
- Skafar et al., Riboflavin metabolism shapes FSP1-driven ferroptosis resistance, accepted at Nature Cell Biology
- Hartmann*, Riedmiller*, Buhr* et al., KIMMDY: A biomolecular reaction emulator, submitted to Nature Communications
- Buhr et al., Collagen’s Triple Helix Reduces the Susceptibility to Base-Catalyzed Hydrolysis, submitted to The Journal of Physical Chemistry B
Force Fields and Topologies

QM/MM simulations reveal mechanistic details of the tetrahedral intermediate formation


More SASA
KIMMDY finds a new radical scavanging candidate in collagen
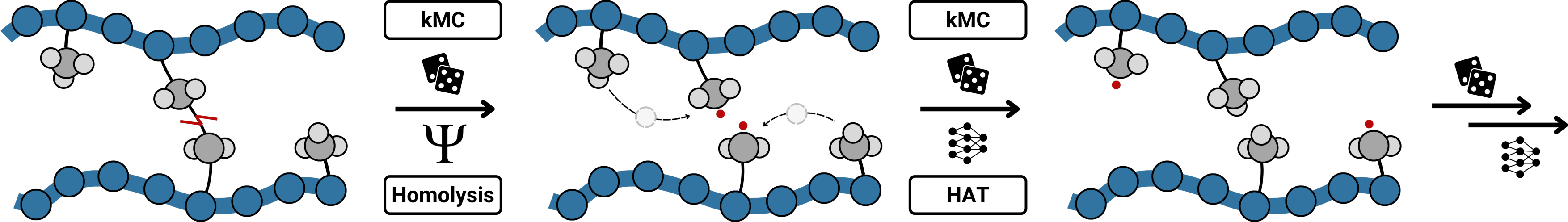
Image sources:
HAT schema by Denis Kieswetter
EPR spectrum and BDEs for DOPA and PYD by Daniel Sucerquia
other BDEs by Wojtek Treyde.
Proton mobility in explicit QM solvent molecules allows breaking of the peptide bond

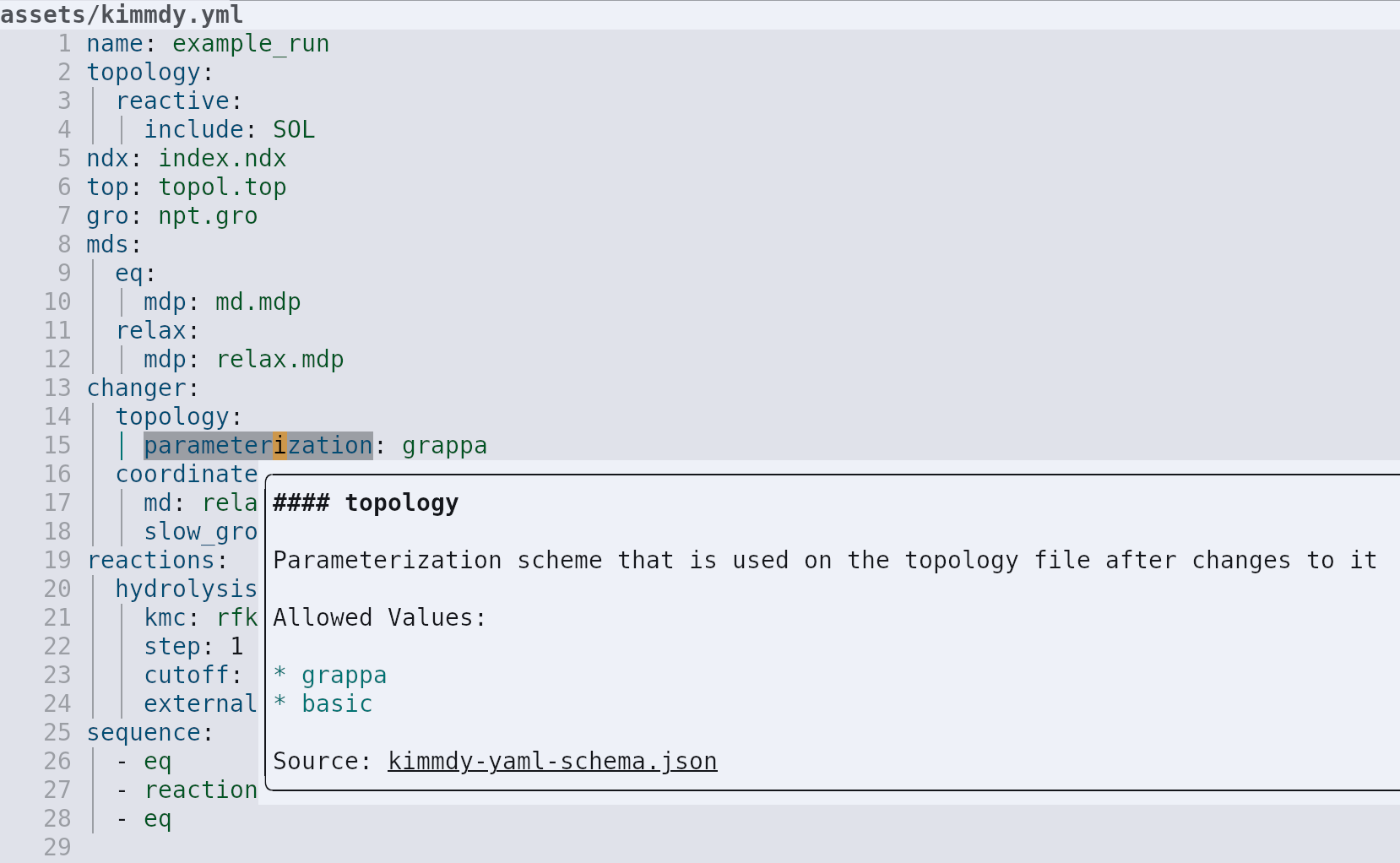
KIMMDY is easy to install and well documented
graeter-group.github.io/kimmdy/

Schema based on a figure by Daniele Procida [13].
KIMMDY is easy to use
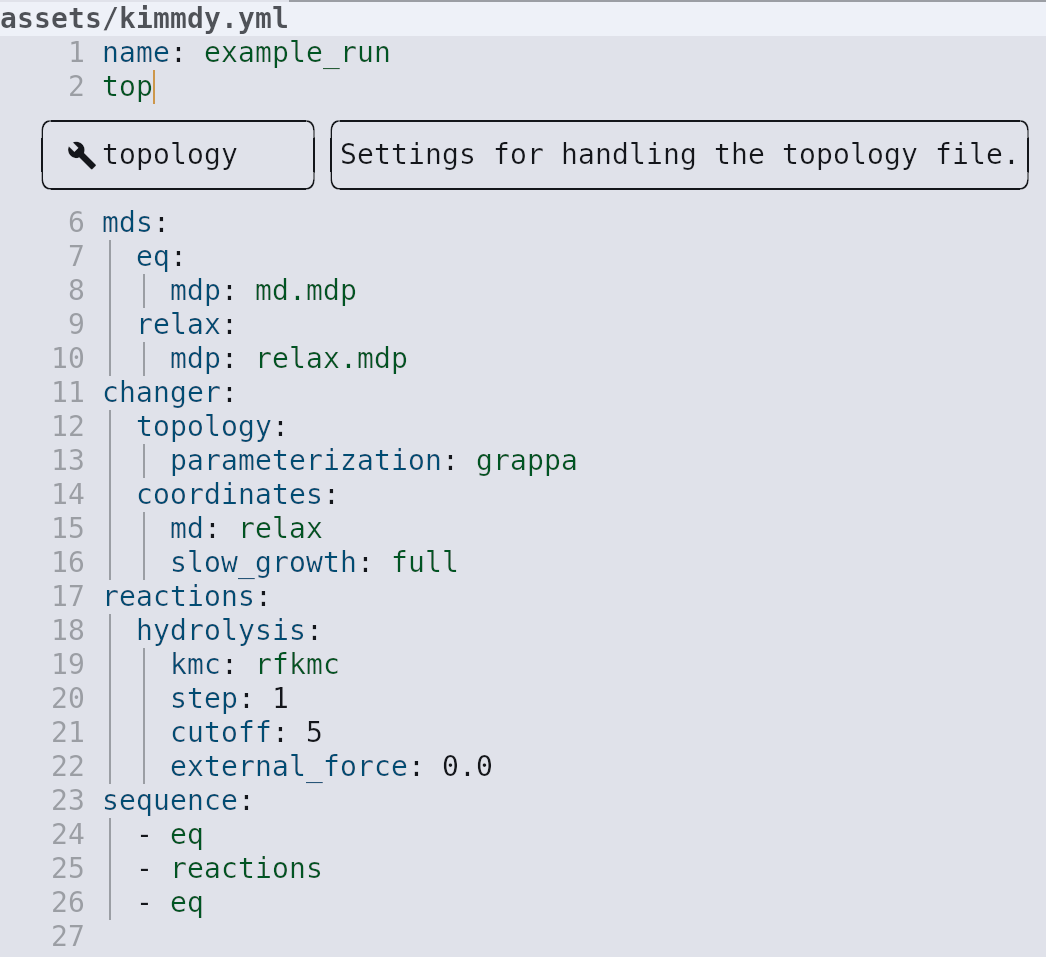

Running KIMMDY: Other cool features
- checkpoints and restarting
- high performance computing support
- extensive testing and logging
- easy to install, use and extend
Direct comparison of hydrolysis and homolysis force response
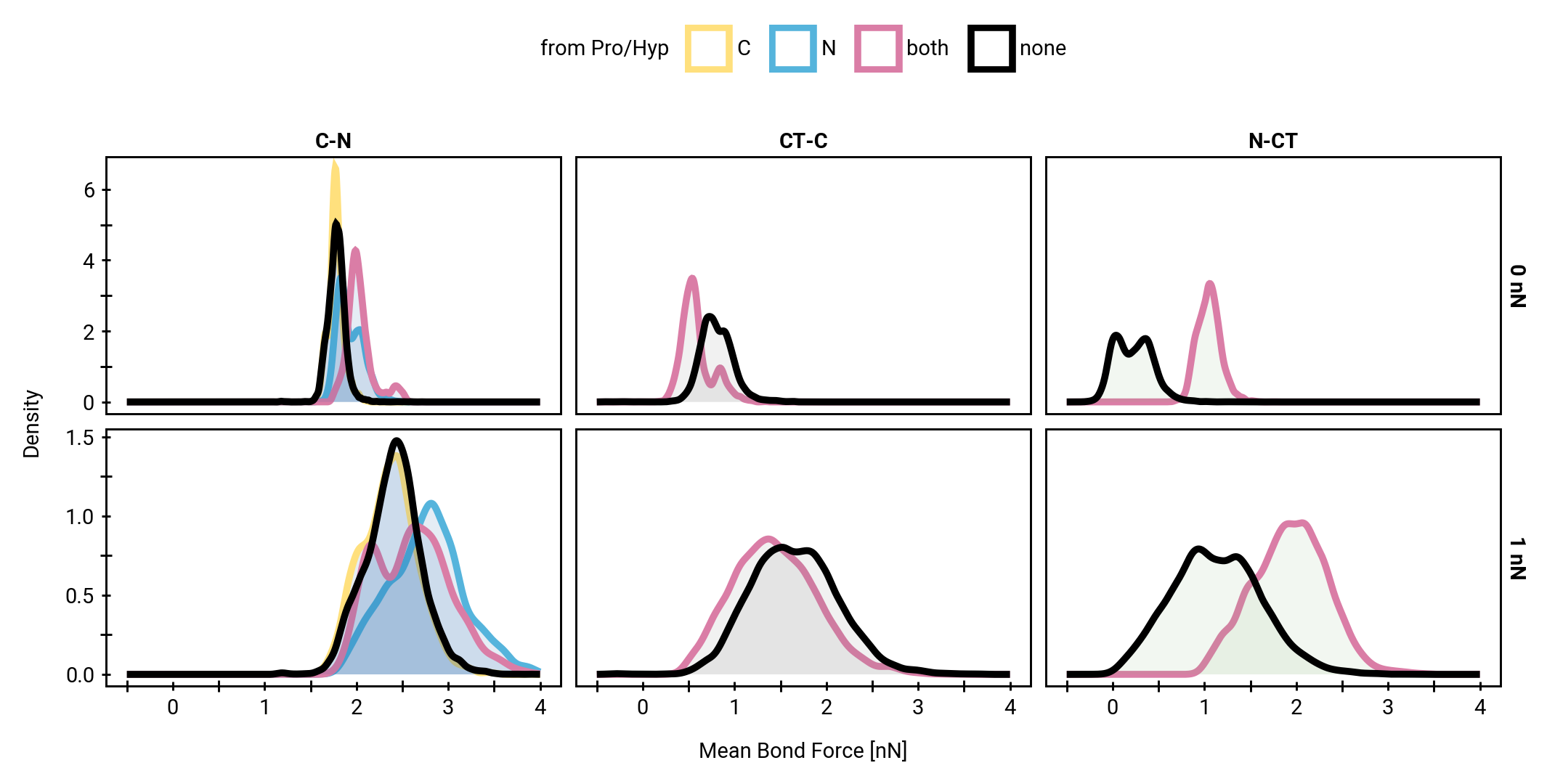
The unique effect of prolines
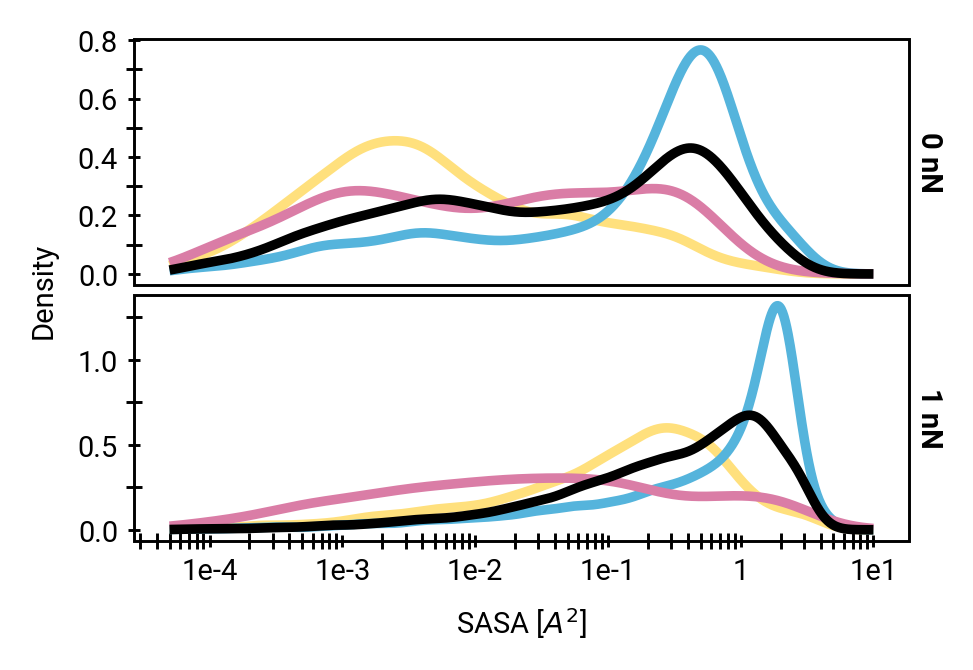
Peptide bonds near the center are still accessible
Supplementary Information
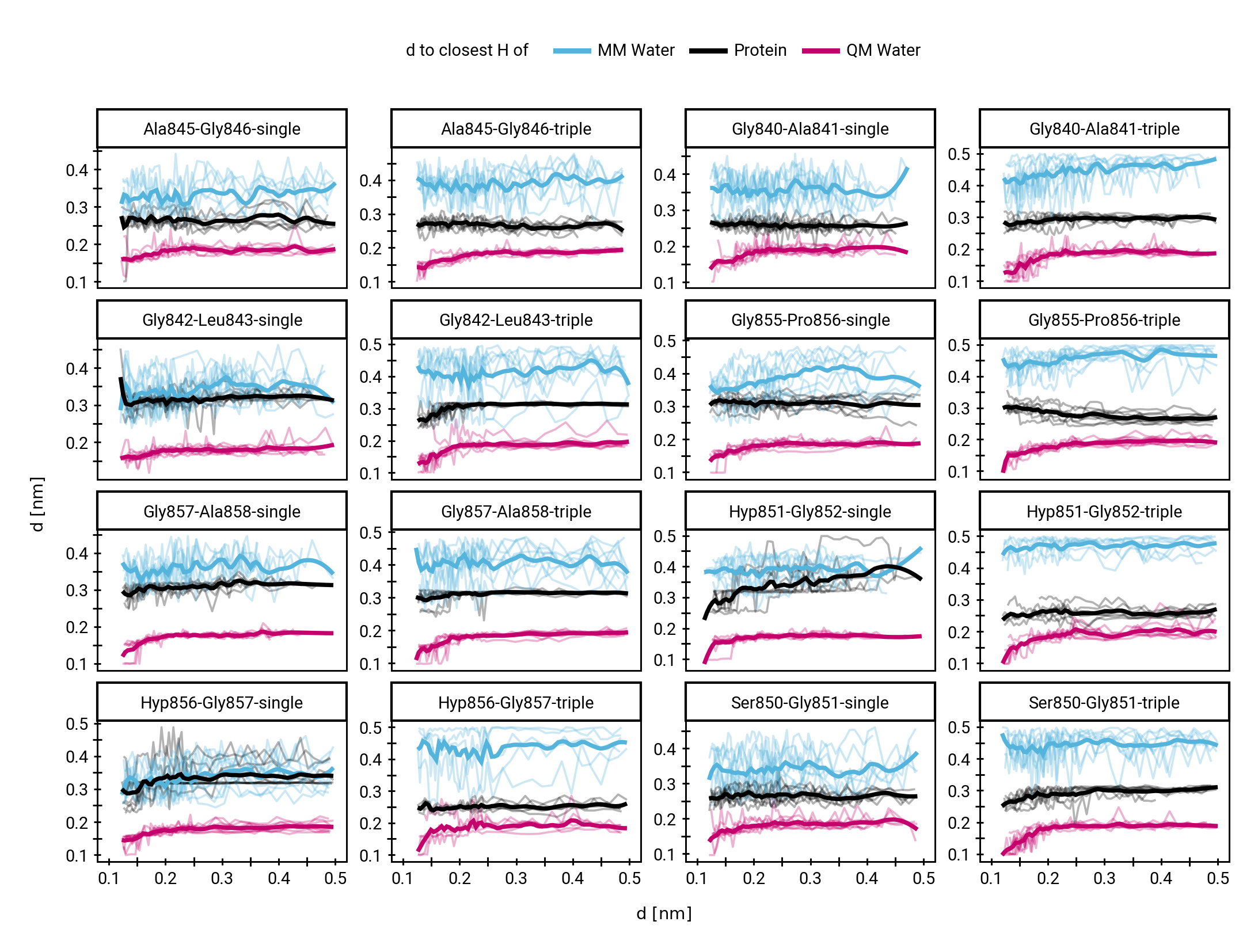
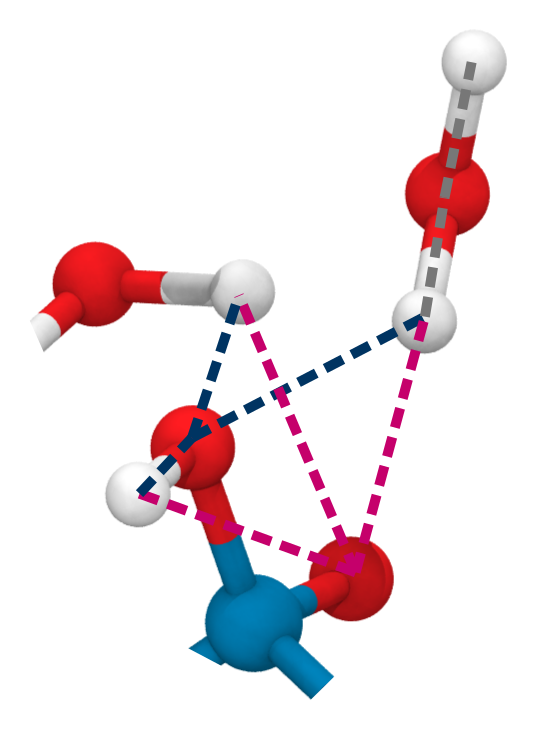
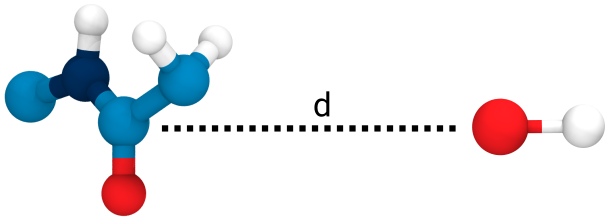
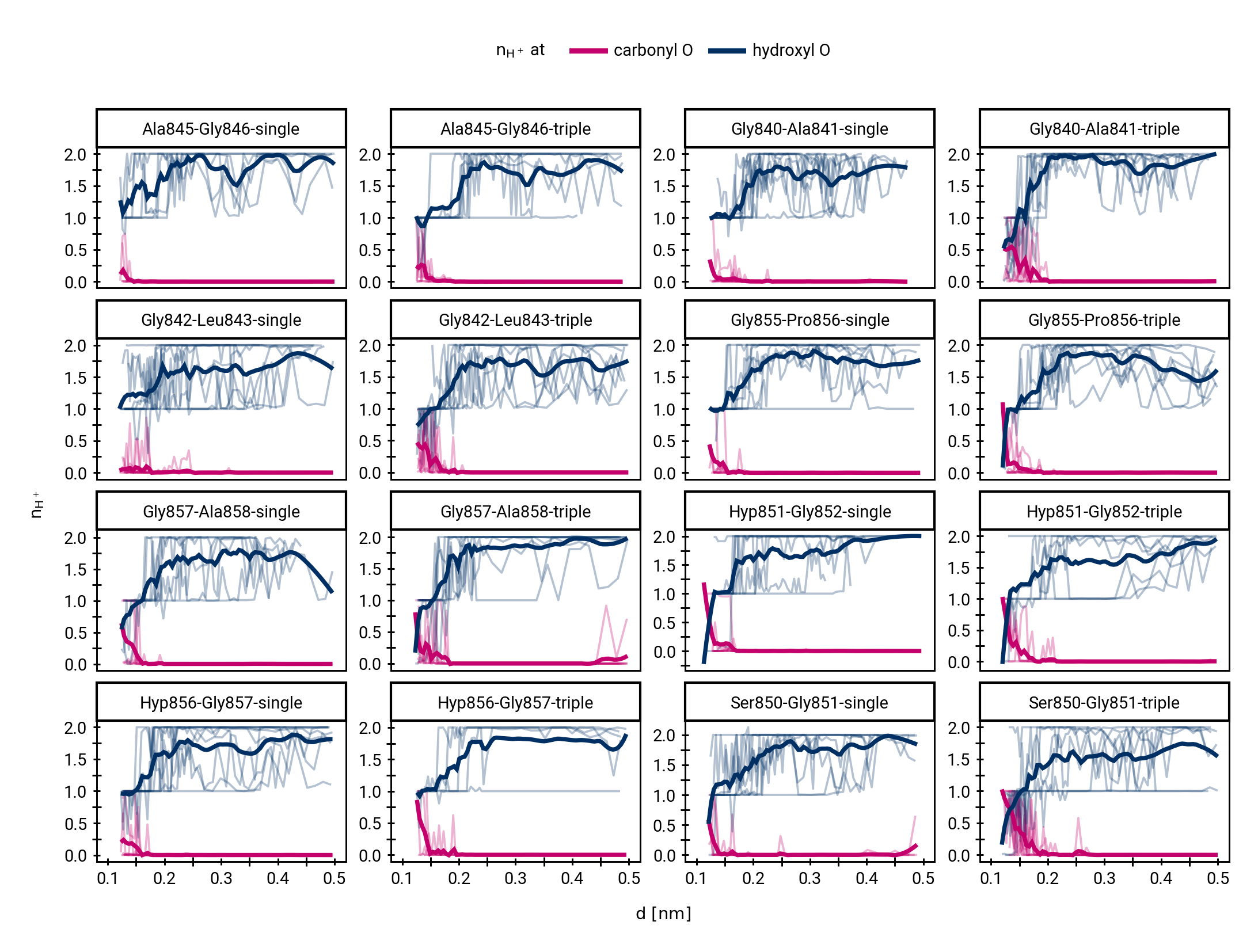
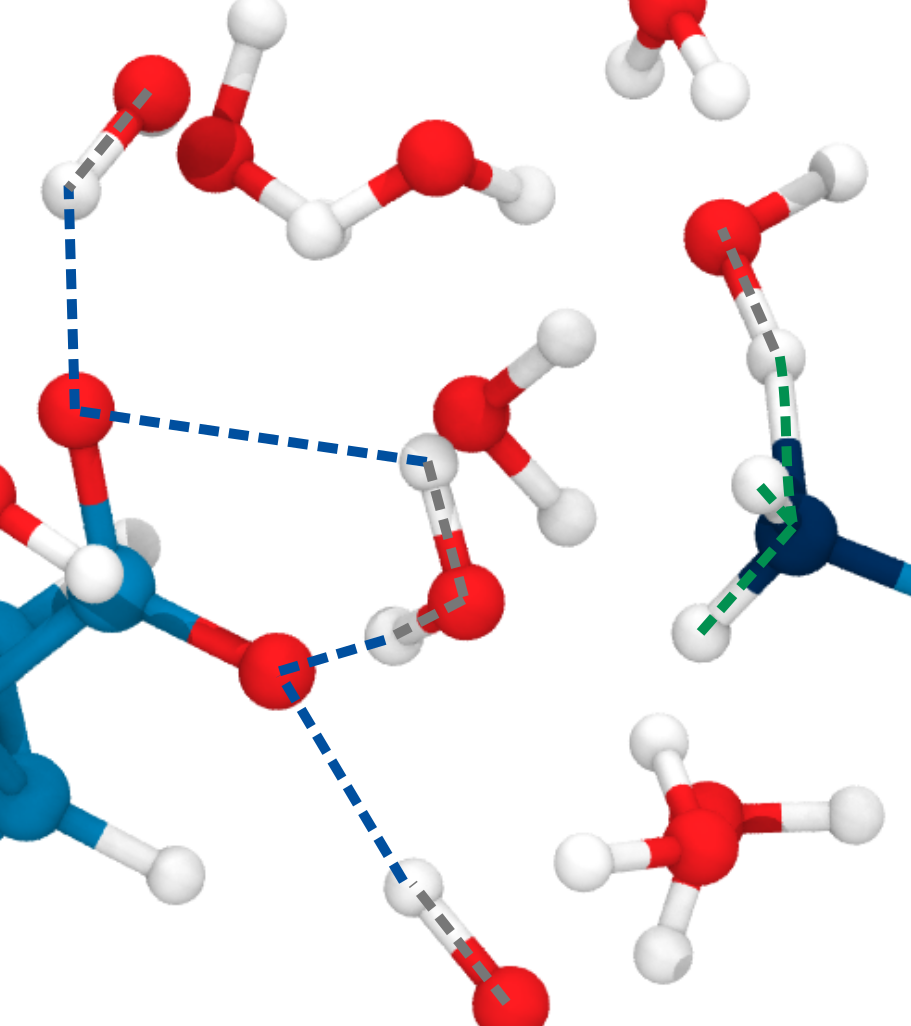
Small multiples plot of the approach of the hydroxide in terms of distances of the carbonyl \(\ce{O}\) of the TI to nearest proton of either a MM solvent molecule (cyan), the protein (black) or the QM water (magenta) across all umbrella windows. Distances are average within each window and the windows belonging to the same approach simulation are connected. The thick lines are a LOESS curve across all windows.
Supplementary Information
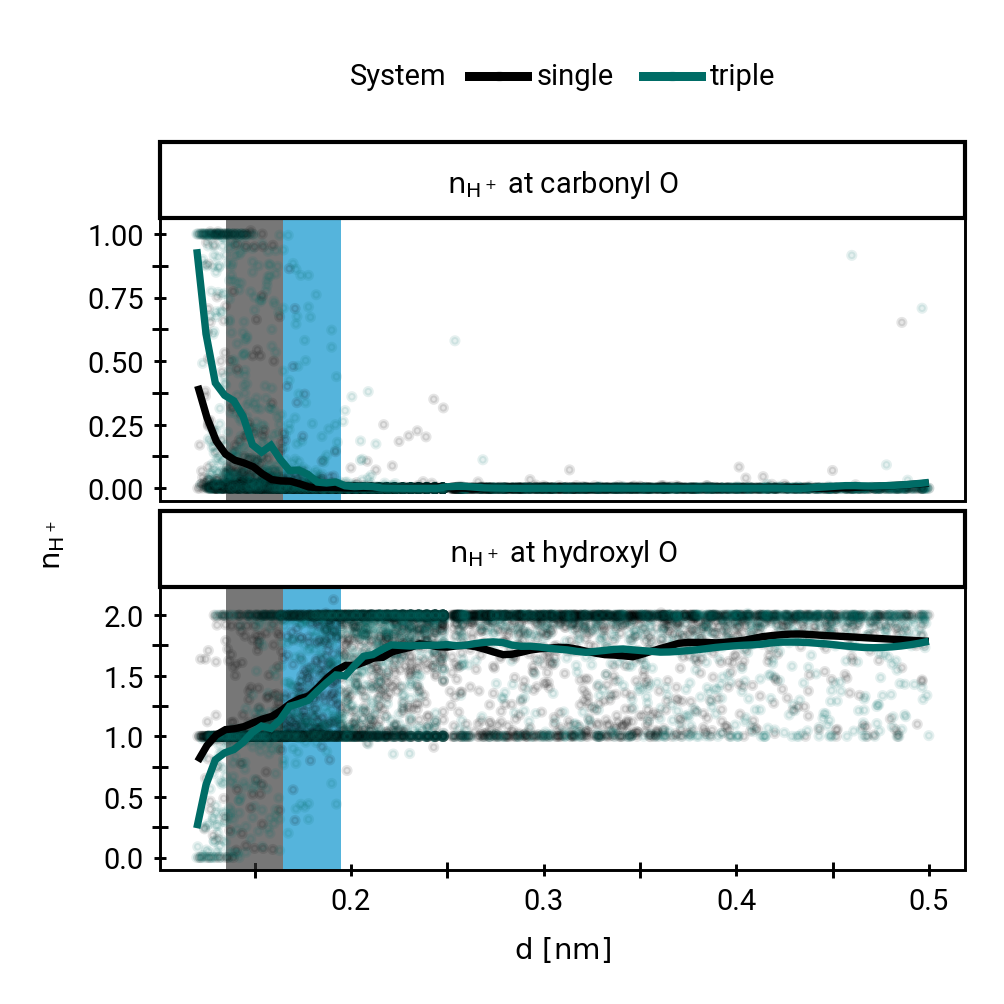
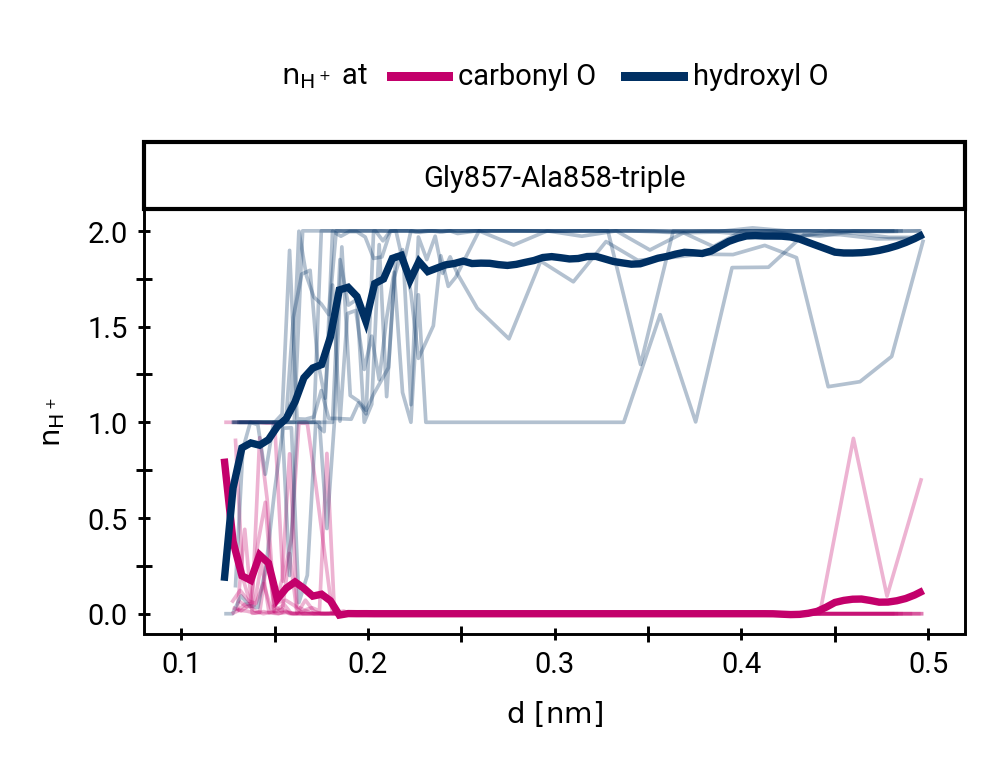
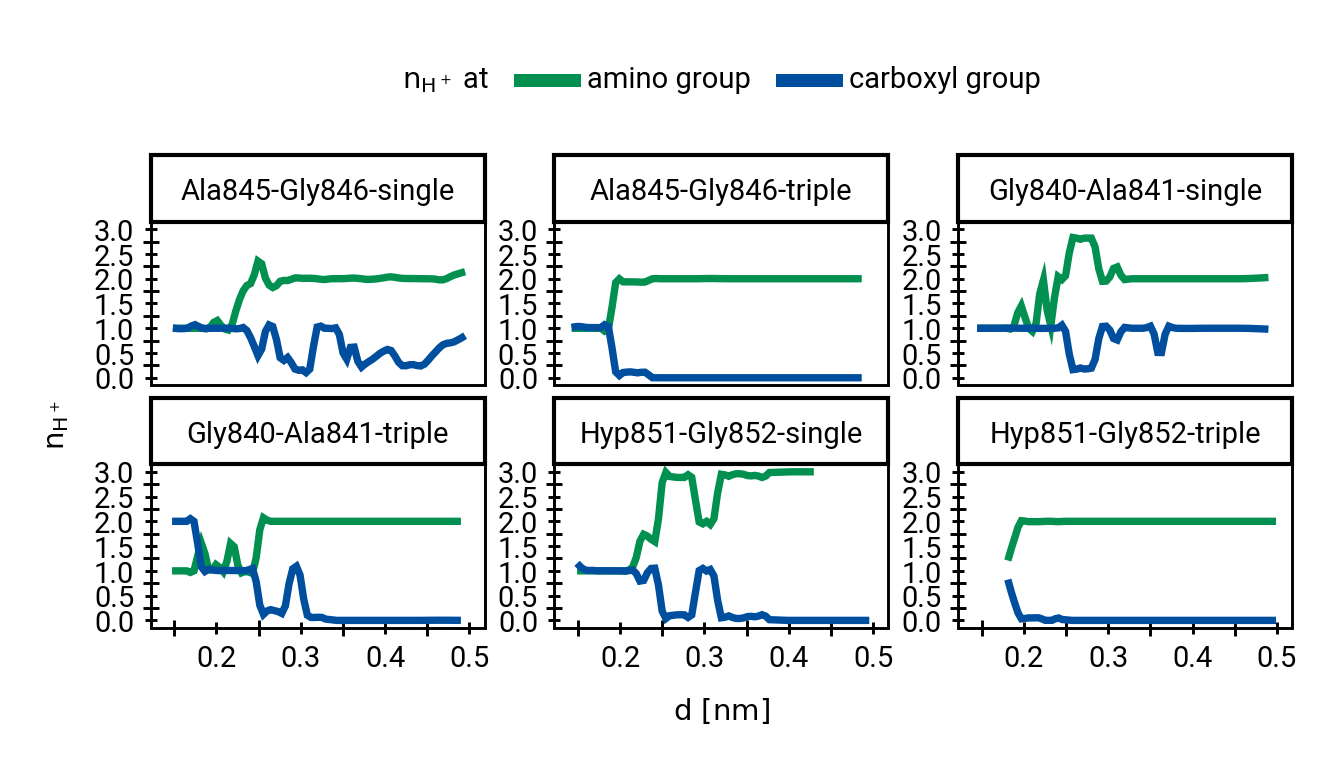
Figure 1: The protonation states are show as the average number of protons assigned to either the hydroxyl \(\ce{O}\) or the carbonyl \(\ce{O}\) with all sampling windows averaged for each system. The region for detection of TS1 is shaded cyan, while the region for detection of the TI is shaded gray.